Abstract
The complete chloroplast genome of Wonwhang (BioSample SAMN05196235), Pyrus pyrifolia, was assembled and analyzed by de novo assembly using whole-genome sequencing data. The accession NC_015996 was used as a reference sequence in this study. The total chloroplast genome size of the Wonwhang was 159,922 bp in length, including a pair of inverted repeat regions (IRs) of 26,392 bp that are separated by a large single-copy region of 72,023 bp and a small single-copy region of 19,235 bp. A total of 132 genes, including 93 protein-coding genes, 31 tRNA genes and eight rRNA genes, were predicted from the chloroplast genomes. Among them, 18 genes occur in IRs, containing nine protein-coding genes, five tRNA genes and four rRNA genes. The GC content of Wonwhang chloroplast genome is 36.6%. The phylogenetic analysis with nine Rosids species and three other species revealed that Wonwhang was clustered with Malus genus.
Pears belong to the family of Rosaceae and have been cultivated in East Asia, Europe and North America for more than 3000 years, and are among the most important fruit crops in temperate regions (Bell Citation1990). Asian pears have a sweet flavour, rich juice and crisp flesh, and more excellent quality and storage-friendly than Occidental pears (Kim Citation2016). In South Korea, pear breeding began in the late 1920s by National Institute of Horticultural and Herbal Science (NIHHS) of Rural Development Administration (RDA). To improve the fruit quality in terms of fruit size, sugar content, flesh firmness and storage quality, many breeders frequently used interspecific hybridization (Kim Citation2016). Consequently, the classification of pears is very difficult because of the occurrence of both natural and artificial interspecific hybrids. Therefore, the genetic relationships and evolutionary history of Asian pears are unclear (Jiang et al. Citation2016). The chloroplast genomes that have a small size, conserved gene content and arrangement, and maternally inheritance are useful resources for evolutionary and phylogenetic studies (Zhang & Sodmergen Citation2010). In this study, we sequenced the complete chloroplast genomes of Wonwhang based on an Illumina platform.
The plant sample of Wonwhang was collected from Naju (latitude: 35° 01′25.6′′N, longitude: 126° 44′38.4′′E), Korea, and representatively identified by Dr. YK Kim of the Pear Research Station, NIHHS, RDA. Total genomic DNA was extracted from young leaves using a DNeasy Plant Mini kit (Qiagen, CA) according to the manufacturer’s instructions. Whole-genome sequencing was performed using an Illumina genome analyzer (NextSeq, Illumina, San Diego, CA) platform at LabGenomics (http://www.labgenomics.co.kr/) in Seongnam-si, Republic of Korea. Genomic libraries with 350-bp insert size were prepared by following the paired-end standard protocol recommended by the manufacturer, and each sample was tagged separately with a different index. De novo assembly was performed using a CLC genome assembler (4.06 beta, CLC Inc., Aarhus, Denmark) with parameters of a minimum 200–600 bp autonomously controlled overlap size, as described by Kim et al. (Citation2015). Low-quality reads were filtered out and the remaining high-quality reads were mapped to the reference chloroplast genome of Housi (Terakami et al. Citation2012) with an average coverage of 80 × on the chloroplast genome. Contig alignment and scaffolding based on paired-end data resulted in a complete circular Wonwhang chloroplast genome sequence. Gene annotation was conducted using CpGAVAS (http://www.herbalgenomics.org/0506/cpgavas/analyzer/annotate). The complete chloroplast genome of Wonwhang was submitted to GenBank under the accession numbers of KX450876.
The complete chloroplast genome of Wonwhang is 159,922 bp in length, with a pair of IR regions of 26,392 bp that separate an LSC region of 72,023 bp and an SSC region of 19,235 bp. It has a GC content of 36.6% and contains 93 protein-coding genes, 31 tRNA genes and 8 rRNA genes. Among them, 18 genes occur in IRs, containing nine protein-coding genes (ndhB, rpl2, rpl23, rps7, rps12, rps19, ycf2 and ycf15(x2)), five tRNA genes (trnI-CAT, trnL-CAA, trnN-GTT, trnR-ACG and trnV-GAC) and four rRNA genes (rrn4.5S, rrn5S, rrn16S and rrn23S). Interestingly, the rps12 gene is triple and the ycf15 gene is quadruple copies in chloroplast genome. Fourteen genes contain one intron and two genes (ycf3 and clpP) contain two introns.
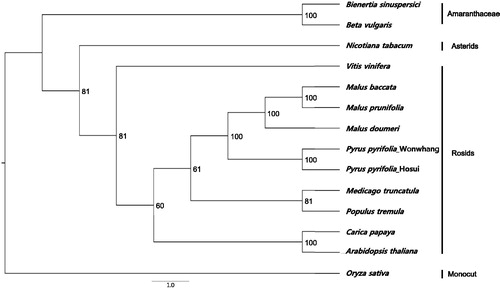
To study its phylogenetic relationships with previously reported complete chloroplast genomes, nine complete chloroplast genome sequences of Rosids and three species from other family were downloaded for analysis. Maximum-likelihood (ML) phylogenetic tree for 13 species with Oryza sativa as an outgroup (). The complete chloroplast genome sequence was aligned using MAFFT v7.222 (Katoh & Standley Citation2013). ML analysis was performed in PHYLIP v3.695 (Felsenstein Citation2013) with support for branches evaluated by 1000 bootstrap replications. The phylogenetic tree showed that Wonwhang chloroplast was clustered with Malus genus and belonged to Rosids clade. The complete chloroplast genome of Wonwhang provides essential and important DNA molecular data for phylogenetic and evolutionary analysis for Asian pears.
Acknowledgements
This work was carried out with the support of the Cooperative Research Program for Agriculture Science & Technology Development (PJ010450) and a grant from the National Academy of Agricultural Science (PJ010880), Rural Development of Administration, South Korea.
Disclosure statement
The authors declare no conflicts of interest.
Additional information
Funding
References
- Bell RL. 1990. Pears (Pyrus). In: Moore JN, Ballington JR Jr, editors. Genetic resources of temperate fruit and nut crops I. Wageningen: International Society for Horticultural Science; p. 655–697.
- Felsenstein J. 2013. Phylip (Phylogeny Inference Package) version 3.695. Seattle: Department of Genome Sciences and Department of Biology, University of Washington.
- Jiang SJ, Zheng X, Yu P, Yue X, Ahmed M, Cai D, Teng Y. 2016. Primitive genepools of Asian pears and their complex hybrid origins inferred from fluorescent sequence-specific amplification polymorphism (SSAP) markers based on LTR retrotransposons. PLoS One. 11:e0149192.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim YK. 2016. Diversity of Pyrus germplasm and breeding of red skin colored and scab resistant pear by using interspecific hybridization [thesis]. Chonnam National University.
- Kim K, Lee SC, Lee J, Lee HO, Joh HJ, Kim NH, Park HS, Yang TJ. 2015. Comprehensive survey of genetic diversity in chloroplast genomes and 45S nrDNAs within Panax ginseng species. PLoS One. 10:e0117159.
- Terakami S, Matsumura Y, Kurita K, Kanamori H, Katayose Y, Yamamoto T, Katayama H. 2012. Complete sequence of the chloroplast genome from pear (Pyrus pyrifolia): genome structure and comparative analysis. Tree Genet Genomes. 8:841–854.
- Zhang Q, Sodmergen. 2010. Why does biparental plastid inheritance revive in angiosperms? J Plant Res. 123:201–206.