Abstract
In this study, we sequenced the complete mitochondrial genome of Thermophis shangrila by using the next-generation sequencing technique. The total length of the mitogenome was 17,407 bp, which was composed of 13 protein coding genes, two rRNA genes (12s and 16s rRNA), 22 tRNA genes, and two control regions (CRI and CRII). The base composition was 32.6% for A, 23.9% for T, 30.0% for C, and 13.5% for G. We added a fragment about 150 bp in length at control region I, which Peng et al. failed to obtain using Sanger dideoxy sequencing.
The hot-spring keel-back (Thermophis), which contains only three species (T. zhaoermii; T. baileyi; T. shangrila), achieves the world’s highest altitude distribution (over 4000 m) among all snakes (Huang et al. Citation2009; He et al. Citation2010; Alex et al. Citation2016). Thermophis shangrila was identified as a new species of Thermophis by Peng et al. (Citation2014). In this study, we sequenced the complete mitochondrial genome of T. shangrila by using the next-generation sequencing (NGS).
T. shangrila were collected from Tianshengqiao Shangri-La, Yunnan province, China. Voucher specimens were deposited at CIB herpetological museum, Chengdu Institute of Biology with the number CIBLJT20150813. The total genomic DNA was extracted from the fresh muscle tissue. We got mitochondrial genome directly from NGS data, which was generated from total genomic DNA extracts (Hahn et al. Citation2013). Then, we downloaded RefSeq (mitogenome of Thermophis zhaoermii) from Organelle Genome Resources database at NCBI (https://www.ncbi.nlm.nih.gov/genome/organelle/) and assembled de novo mitogenome using MITObim (Paszkiewicz & Studholme Citation2010). Gene structure was predicted by Mitos (Bernt et al. Citation2013). The RefSeq was also used to correct the mitogenomic structure manually. The base composition was calculated by MEGA5 (Tamura et al. Citation2001). The tRNA genes were scanned by tRNAscan-SE (Lowe & Eddy Citation1997).
The complete mitogenome sequence together with gene annotations were deposited in the GenBank under the accession number MF066951. The total length of the T. shangrila mitochondrial genome was 17,407 bp, which was slightly longer than 17,327 bp as reported by Peng et al. (Citation2016). The base composition was 32.6% for A, 23.9% for T, 30.0% for C, and 13.5% for G. The mitogenome we sequenced contained 13 protein coding genes, two rRNA genes (12s and 16s rRNA), 22 tRNA genes, and two control regions (CRI and CRII), which were similar to the other reported snakes (He et al. Citation2010). As for sequence features of protein-coding genes, start codons were ATG for COXII, COXIII, ATP8, ATP6, ND4L, ND4, ND5, and CYTB; ATA for ND1, ND2, and ND3; and GTG for the remaining one (COXI). TAA was the stop codon for ATP8, ATP6, ND4L, and ND5. AGG was the stop codon for ND4. Furthermore, the length of 22 tRNA genes ranged from 57 to 74 bp, of which the shortest one was tRNASer (AGY) and the longest was tRNAAsn. The sequence length of 12s and 16s rRNA were 925 bp and 1479 bp, respectively, which were separated by tRNAVal. As reported by Peng et al. (Citation2016), CRI was 1175 bp, which was longer than CRII, as CRII was 1026 bp.
For convincing the mitochondrial DNA sequences, a maximum likelihood (ML) phylogenetic tree was re-constructed in the program RAxML-HPC (Stamatakis Citation2006) using alignment of coding regions of 11 mitochondrial genomes of Colubridae (). Phylogenetic analysis showed that T. shangrila was located within the lineage of Colubridae and was sister species of T. zhaoermii as previously described (Sun et al. Citation2011; Alex et al. Citation2016).
Figure 1. The ML tree of 11 species from Colubridae was constructed based on the protein-coding genes except ND6. The GenBank accession number is as follows: Dinodon rufozonatum (KF148622), Rhabdophis tigrinus (NC030210), Elaphe schrenckii (NC027605), Elaphe anomala (NC027001), Elaphe schrenckii (NC027605), Cyclophiops major (NC028048), Ptyas dhumnades (KF148621), Oligodon ningshaanensis (KJ719252), Hypsiglena jani texana (EU728592), Hypsiglena slevini (EU728584), Thermophis zhaoermii (GQ166168).
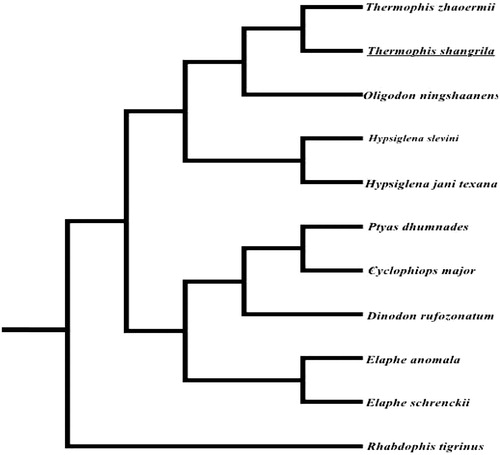
In this study, we revealed the complete mitochondrial sequence of T. shangrila and sequenced the fragment from 3809 to 3961 bp, which failed to be shown by Peng et al. (Citation2016). We hope that this determination about T. shangrila mitochondrial genome will provide useful resources for better understanding the evolution of genus Thermophis and the whole Colubridae.
Acknowledgements
We thank Jin-Long Ren for assistance in sample collection.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alex F, Mckelvy AD, Lee GL, Bell CD, Lailvaux SP. 2016. A species-level phylogeny of extant snakes with description of a new colubrid subfamily and genus. PLoS One. 11:1–31.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucl Acids Res. 41:e129.
- He M, Feng JC, Zhao EM. 2010. The complete mitochondrial genome of the Sichuan hot-spring keel-back (Thermophis zhaoermii; Serpentes: Colubridae) and a mitogenomic phylogeny of the snakes. Mitochondrial DNA. 21:8–18.
- Huang S, Liu SY, Guo P, Zhang YP, Zhao EM. 2009. What are the closest relatives of the hot-spring snakes (Colubridae, Thermophis), the relict species endemic to the Tibetan Plateau? Mol Phylogenet Evol. 51:438–446.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 25:955–964.
- Paszkiewicz K, Studholme DJ. 2010. De novo assembly of short sequence reads. Brief Bioinformat. 11:457–472.
- Peng L, Changhu LU, Huang S, Guo P, Zhang Y. 2014. A new species of the genus Thermophis (Serpentes: Colubridae) from Shangri-La, Northern Yunnan, China, with a proposal for an eclectic rule for species delimitation. Asian Herpetol Res. 5:228–239.
- Peng LF, Weng SY, Yang DC, Lu CH, Huang S. 2016. Complete mitochondrial genome of the Xianggelila Hot-spring snake, Thermophis shangrila (Reptilia, Colubridae). Mitochondrial DNA Part B Resources. 1:536–537.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Sun X, Liu S, Huang S. 2011. Tibetan plateau relict snakes of the genus Thermophis and their relationship to new world relict snakes. Asian Herpetol Res. 2:161–168.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2001. MEGA5:molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
