Abstract
The complete mitochondrial genome of the microalgae Dunaliella salina strain SQ (GenBank accession number: KX641170) was de novo assembled and annotated using Illumina MiSeq sequencing data. The mitochondrial genome is 41,904 bp long with 31.85% GC content and contains 7 protein-coding genes, 16 introns, 3 ribosomal RNA genes and 3 transfer RNA genes. To date, only two complete mitochondrial genomes of Dunaliella salina strains have been reported, and this genome provides knowledge to the study of genetic variations and evolution of mitochondrial genomes of Dunaliella salina strains.
Green microalgae biomass has been used for decades; in recent years, secondary metabolites extracted from green microalgae have gained interest for their commercial and biotechnological potential. The unicellular microalgae Dunaliella salina lives in hypersaline environments and under stress conditions can produce about 10% β-carotene of biomass dry weight (Lamers et al. Citation2008). β-carotene is an important pro-vitamin A, and vitamin A is essential to human body for maintaining normal vision, tissue differentiation and immune competence (Weber & Grune Citation2012; Sommer & Vyas Citation2012). To date, only two complete mitochondrial genomes of D. salina strains have been reported: D. salina CCAP 19/18 (Smith et al. Citation2010) and D. salina CONC-001 (Del Vasto et al. Citation2015); these genomes present several differences in genome size and GC content between strains. Here we report the complete mitochondrial genome of D. salina SQ to provide information about the genome architecture between D. salina strains.
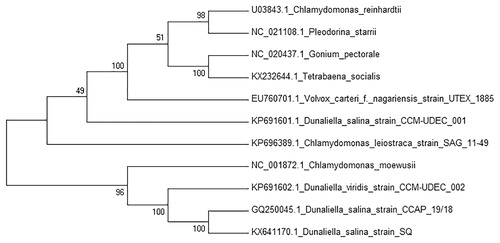
Dunaliella salina was collected from a hypersaline lagoon in San Quintin, Baja California, México (30° 32’ 13.91" N, 116° 2’ 1.22" W). A single cell was isolated and cultured in modified liquid medium (Feng et al. Citation2009) with 250 mM NaCl. Mitochondrial DNA was extracted with AxyPrep Multisource Genomic DNA Miniprep Kit (Axygen Biosciences, Union City, CA), and mtDNA was sequenced using Illumina MiSeq (UC, San Diego, CA) obtaining 300-bp paired-end reads. The genome was de novo assembled using A5-miseq pipeline (Coil et al. Citation2014) and Ray v2.3.2 (Boisvert et al. Citation2010). Annotation of the mitochondrial genome was performed with RNAweseal (Lang et al. Citation2007), MFannot (Beck & Lang Citation2010) and manual curated, and results were validated using BLAST searches. The annotated genome has been deposited in the GenBank database under accession number KX641170. Whole mitochondrial genome sequence of D. salina has a circular structure of 41,904 bp, containing 7 protein-coding genes, 16 introns, 3 ribosomal RNA genes and 3 transfer RNA genes. The contents of A, G, T and C are 32.6%, 16.7%, 35.5% and 15.2%, respectively, with 31.85% of GC content. All protein-coding genes started with ATG and ended by TAA as a stop codon, except nad6 which ended by TGA. The nad1 gene contains 1 intron and 1 intronic open reading frame GIY-YIG homing endonuclease, nad4 1 intron, cob gene 5 introns and 1 intronic open reading frame LAGLIDADG homing endonuclease, cox1 gene 7 introns and 2 intronic open reading frames LAGLIDADG homing endonuclease, nad5 gene 2 introns and 1 open reading frame GIY-YIG homing endonuclease. For the phylogenetic analysis, 10 mitochondrial genome sequences from other microalgae were selected and downloaded from NCBI database and a neighbour-joining tree with 500 bootstraps was constructed using MEGA7 (Kumar et al. Citation2016) ().
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Beck N, Lang B. 2010. MFannot, organelle genome annotation webserver [Internet]. Available from: http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl.
- Boisvert S, Laviolette F, Corbeil J. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 17:1519–1533.
- Coil D, Jospin G, Darling AE. 2014. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31:587–589.
- Del Vasto M, Figueroa-Martinez F, Featherston J, Gonzalez MA, Reyes-Prieto A, Durand PM, Smith DR. 2015. Massive and widespread organelle genomic expansion in the green algal genus Dunaliella. Genome Biol Evol. 7:656–663.
- Feng S, Xue L, Liu H, Lu P. 2009. Improvement of efficiency of genetic transformation for Dunaliella salina by glass beads method. Mol Biol Rep. 36:1433–1439.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lamers PP, Janssen M, De Vos RC, Bino RJ, Wijffels RH. 2008. Exploring and exploiting carotenoid accumulation in Dunaliella salina for cell factory applications. Trends Biotechnol. 26:631–638.
- Lang BF, Laforest MJ, Burger G. 2007. Mitochondrial introns: a critical view. Trends Genet. 23:119–125.
- Smith DR, Lee RW, Cushman JC, Magnuson JK, Tran D, Polle JE. 2010. The Dunaliella salina organelle genomes: large sequences, inflated with intronic and intergenic DNA. BMC Plant Biol. 10:83.
- Sommer A, Vyas KS. 2012. A global clinical view on vitamin A and carotenoids. Am J Clin Nutr. 96:1204S–1206S.
- Weber D, Grune T. 2012. The contribution of ß-carotene to vitamin A supply of humans. Mol Nutr Food Res. 56:251–258.