Abstract
The mitochondrion genome of Occidentarius platypogon was assembled from Illumina short reads, and consisted of 16,714 base pairs, with 13 protein-coding, two ribosomal RNAs (rRNAs), and 22 transfer RNA (tRNA) genes. Base composition is 30.7% A, 26.4% T, 28.5% C, and 14.4% G, and 42.9% GC content. Two start codon (ATG and GTG) and seven stop codon (TAA, ACT, CCT, TTA, CAT, AAT, and TAG) patterns were found in protein-coding genes. Control region presented the highest A + T (64%) and lowest G + C content (35.7%) among all mitochondrial regions.
The cominate catfish, Occidentarius platypogon, belongs to a monotypic genus which include only this species, and its distribution is limited to the Eastern Pacific from Mexico to Peru (Betancur-R et al. Citation2007). This species is of relevant interest for artisanal fishing along the Sinaloa coast (northwest of Mexico), and is also incidentally caught by industrial and artisanal shrimp fisheries (Amezcua et al. Citation2006; Madrid-Vera et al. Citation2007; Amezcua & Muro-Torres Citation2012). In spite of abundance and economic importance of this species, there are few studies that describe their biology, ecology and genetics. In Mazatlán coast, O. platypogon present a synchronic gonad development and a spawning season from May to August, with males incubating the eggs in the oral cavity (Amezcua & Muro-Torres Citation2012).
To determine the complete mitochondrial genome of O. platypogon, we collected a specimen captured in Sinaloa, Mexico (23°28′32.5″N–106°37′28.2″W). DNA was extracted from fresh muscle tissue using the Wizard® Genomic DNA Purification kit (Promega, Madison, WI). A genomic DNA library was constructed with the Kapa gDNA library preparation kit (Kapa Biosystems, Wilmington, MA) using multiplex index, and the library was then sequenced alongside other barcoded libraries using a single lane (2 × 125 paired-end reads) in a MiSeq platform (Illumina, San Diego, CA). After demultiplexing, reads were pre-processed using Trimmomatic 0.33 (Bolger et al. Citation2014) for trim low-quality ends (Q score <20), residual adapters and remove reads shorter than 100 bases. All reads were analysed for quality control (QC) with FastQC v0.10.1 (Babraham Institute, Cambridge, UK) (Andrews Citation2011) before and after quality trimming. 21’391,977 pair-end reads were finally obtained.
Using the complete mitochondrion genome of Netuma thalassina (GenBank accession number: KU986659.1) as a reference, the complete genome was obtained by multi-step assembly (7 iterations) using the MITObim ver.1.7 (Hahn et al. Citation2013). The mitogenome of O. platypogon (GenBank accession number KY930717) has a length of 16,714 bp and a base composition of A 30.7%, T 26.4%, C 28.5%, and G 14.4%, and the GC content of 42.9%. The complete mitochondrial genome contains all typical genes found in most vertebrate mitogenomes: 13 protein-coding genes, 22 transference RNA genes, two ribosomal RNAs, and one control region or d-loop (). Protein coding genes initiate by the typical ATG codon, except for the COX1 gene, which presented GTG as start codon. Almost all genes presented TAA or ACT as stop codon, the rest used a different one (). D-loop region was 1123 bp, presenting the highest A + T content of 64.3% among all mitochondrial regions. No evidence of SNP variation over a frequency of 0.05 was found.
Figure 1. Maximum-likelihood (ML) phylogenetic tree of Occidentarius platypogon and the other 11 species of 8 families using Clarias fuscus and Heteropneustes fossilis as an outgroup. Number above each node indicates the ML bootstrap support values. In parenthesis the access numbers from NCBI database.
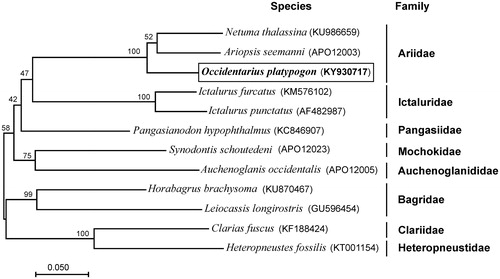
Table 1. Annotation of the complete mitochondrial genome of Occidentarius platypogon.
We validated the phylogenetic position of O. platypogon with a maximum-likelihood tree (500 boostrap replicates) of complete mtDNA from the other 11 species using MEGA6 (Tamura et al. Citation2013). The phylogenetic position of O. platypogon was closely clustered with two species of Ariidae family (). The newly determined mitogenome will help to understand the evolution of Ariidae family.
Acknowledgements
We are grateful to Quetzalli Yasú Abadia-Chanona for help with figure composition. Thanks to Paul Mendivil and Juan Antonio Maldonado for their help in field and lab. NCSS and RLH were benefited for an economic support through the program Cátedras CONACYT (No.: 2137 and 3285 respectively).
Disclosure statement
The authors report no conflicts of interest. They alone are responsible for the content and writing of the manuscript.
Additional information
Funding
References
- Amezcua F, Madrid-Vera J, Aguirre-Villaseñor H. 2006. Effect of the artisanal shrimp fishery on the ichthyofauna in the coastal lagoon of Santa Maria la Reforma, Gulf of California. Cienc Mar. 32:97–109.
- Amezcua F, Muro-Torres V. 2012. Reproductive biology of the cominate sea catfish Occidentarius platypogon (Pisces: Ariidae) from the southeastern Gulf of California. Lat Am J Aquat Res. 40:428–434.
- Andrews S. 2011. FastQC high throughput sequence QC report v.0.10.1. Babraham Bioinformatics.
- Betancur-R R, Arturo AP, Bermingham E, Cooke R. 2007. Systematics and biogeography of New World sea catfishes (Siluriformes: Ariidae) as inferred from mitochondrial, nuclear, and morphological evidence. Mol Phylogenet Evol. 45:339–357.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics. 30:2114–2120.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Madrid-Vera J, Amezcua F, Morales-Bojorquez E. 2007. An assessment approach to estimate biomass of fish communities from bycatch data in a tropical shrimp-trawl fishery. Fish. Res. 1:81–89.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
