Abstract
The chihuil sea catfish (Bagre panamensis) is endemic of the Eastern Pacific and is a species of fishery importance in the Mexican Pacific. The complete mitochondrial genome of Bagre panamensis has been assembled from Illumina sequencing data. The circular genome was 16,714 bp in lengh, and consist of 13 protein-coding, two ribosomal RNAs (rRNAs), and 22 transfer RNA (tRNA) genes. Base composition is 30.8% A, 26.6% T, 28.2% C, and 14.4% G, and 42.6% GC content. Protein-coding genes present two start codon (ATG and GTG) and eight stop codon (TAA, TCT, CCT, TTA, CAT, AAT, ATT, and TAG). The control region possesses the highest A + T (64.4%) and lowest G + C content (35.6%) among all mitochondrial regions. These data would contribute to the evolutionary studies of related taxa.
The Ariidae familie (Order: Siluriformes) is widely distributed in world tropical shelves, includes 150–200 species approx. of which one third is endemic to American coasts (Betancur-R et al. Citation2007; Marceniuk & Menezes Citation2007). The Ariidae familie present a monophyletic ancestry, well-supported by morphological and molecular traits (Diogo Citation2004; Kailola Citation2004; Sullivan et al. Citation2006; Betancur-R et al. Citation2007); however, the systematics of its species is complex and there are many problems on its nomenclatural (Marceniuk & Menezes Citation2007). The ariids are predominantly marine species of which the chihuil catfish (Bagre panamensis) is endemic of the Eastern Pacific, it is distributed from southern California to northern Peru including Gulf of California and Galapagos Islands (Cooke Citation1992; Allen & Robertson Citation1994). Bagre panamensis is a demersal fish that habitat in muddy bottoms near to shore (177 m depth approx.), estuaries and mangroves (Cooke Citation1992).
In this study, we determined the complete mitochondrial genome of B. panamensis for first time. One specimen was collected from artisanal fishery Sinaloa, Mexico (23°28′32.5″N - 106°37′28.2″W). DNA was extracted from fresh muscle tissue using the Wizard® Genomic DNA Purification kit (Promega, Madison, WI). A genomic DNA library was constructed with the Kapa DNA library preparation kit (Kapa Biosystems, Wilmington, MA) using multiplex index, and the library was then sequenced alongside other barcoded libraries using a single lane (2 × 125 paired-end reads) in a MiSeq platform (Illumina, San Diego, CA). Reads were pre-processed using Trimmomatic v0.33 (Bolger et al. Citation2014) for trim low-quality ends (Q score <20), residual adapters and remove reads shorter than 100 bases. The obtained sequences were demultiplexed, and the recovered reads were analysed for quality control with FastQC v0.10.1 (Babraham Institute, Cambridge, UK) (Andrews Citation2011). 21′391,977 pair of high-quality reads (Q score >25) were recovered. The complete mitochondrion genome was obtained using MITObim v1.7 (Hahn et al. Citation2013), using the giant catfish mitogenome Netuma thalassina (GenBank accession number: KU986659.1) as a reference. The final assembly was annotated using MitoAnnotator pipeline (Iwasaki et al. Citation2013).
The mitogenome of B. panamensis (GenBank accession number KY930718) has a length of 16,673 bp with a base composition of A 38.8%, T 26.6%, C 28.2%, and G 14.4% (42.6% of GC content). The mitogenome contains all typical genes of vertebrate: 13 protein-coding genes, 22 transference RNA genes, two ribosomal RNAs, and one control region or d-loop (). Almost all protein-coding genes initiated by typical ATG codon, except for the COX1 gene initiated by the GTC. For the stop codon, almost all genes presented TAA or CCT, the rest used a different one (). The control region (D-loop) is 1080 bp in length which was located between the tRNA-Pro and tRNA-Phe; it had the highest A + T content of 64.4% and lowest G + C content (35%) among all mitochondrial regions.
Figure 1. Maximum-likelihood (ML) phylogenetic tree of Bagre panamensis and the other 11 species of 8 families using Clarias fuscus and Heteropneustes fossilis as an outgroup. Number above each node indicates the ML bootstrap support values. In parenthesis the access numbers from NCBI database.
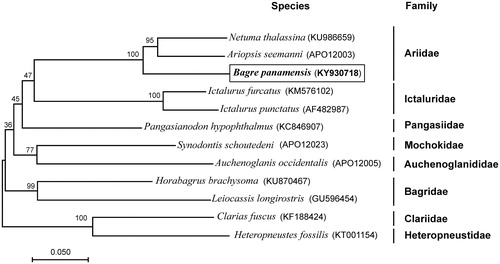
Table 1. Annotation of the complete mitochondrial genome of Bagre panamensis.
To validate the phylogenetic position of B. panamensis, we used MEGA6 (Tamura et al. Citation2013) to construct a maximum-likelihood tree (500 boostrap replicates) containing complete mtDNA of the other 11 species (). The phylogenetic position of B. panamensis was closely clustered with Netuma thalassina and Ariopsis seemanni, the three species belong to Ariidae family which are considered new world catfishes.
Acknowledgments
We are grateful to Paul Mendivil and Juan Antonio Maldonado for their help in field and lab. NCSS and RLH were benefited for an economic support through the program Cátedras CONACYT (No.: 2137 and 3285, respectively).
Disclosure statement
The authors report no conflicts of interest. They alone are responsible for the content and writing of the manuscript.
Additional information
Funding
References
- Allen GR, Robertson DR. 1994. Fishes of the tropical eastern Pacific. Honolulu, USA: University of Hawaii Press.
- Andrews S. 2011. FastQC high throughput sequence QC report v.0.10.1. Babraham Bioinformatics.
- Betancur-R R, Arturo AP, Bermingham E, Cooke R. 2007. Systematics and biogeography of New World sea catfishes (Siluriformes: Ariidae) as inferred from mitochondrial, nuclear, and morphological evidence. Mol Phylogenet Evol. 45:339–357.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 30:2114–2120.
- Cooke R. 1992. Prehistoric nearshore and littoral fishing in the eastern tropical Pacific: an ichthyological evaluation. J World Prehist. 6:1–49.
- Diogo R. 2004. Muscles versus bones: catfishes as a case study for a discussion on the relative contribution of myological and osteological features in phylogenetic reconstructions. Anim Biol. 54:373–391.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Mabuchi K, Takeshima H, Miya M, Nishida M. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Kailola PJ. 2004. Phylogenetic Exploration of the Catfish Family Ariidae (Otophysi: Siluriformes). Beagle: Records of the Museums and Art galleries of the Northem territory. 87–166p.
- Marceniuk AP, Menezes NA. 2007. Systematics of the family Ariidae (Ostariophysi, Siluriformes), with a redefinition of the genera. Zootaxa. 1416:1–126.
- Sullivan JP, Lundberg JG, Hardman M. 2006. A phylogenetic analysis of the major groups of catfishes (Teleostei: Siluriformes) using rag1 and rag2 nuclear gene sequences. Mol Phylogenet Evol. 41:636–662.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
