Abstract
In this study, we firstly reported the complete mitochondrial genome of Copadichromis trimaculatus. The whole mitochondrial genome is 16,581 bp in length, including 2 ribosomal RNA genes, 22 transfer RNA genes and 13 protein-coding genes. Its GC content is 46.00%, similar to Copadichromis virginalis (45.74%). We also analyzed its phylogenic relationship with other 14 related species, which would facilitate our understanding of the phylogenic relationship of Cichlidae mitochondrial genome.
In this study, we firstly reported the complete mitochondrial genome of C. trimaculatus (Genbank accession: MF033352), which would help understand the phylogenic relationship of the Cichlidae family. The C. trimaculatus sample used in this study was collected from the Southeast Arm of Lake Malawi and sequenced by the Wellcome Trust Sanger Institute (SC) in their Cichlid diversity sequencing WTMGM student project. We used SOAPaligner/soap2 (V2.21) (Li et al. Citation2009) to map all the raw reads of the whole genome of a C. trimaculatus (SRA: ERP002088) to the reference mitochondrial genome, Copadichromis virginalis (Genbank accession: NC_029761). To get the complete mitochondrial genome of C. trimaculatus, we assembled these reads which could map to the reference genome by SPAdes3 (V3.1.0) (Bankevich et al. Citation2012). Moreover, DOGMA (Wyman et al. Citation2004) and tRNAscan-SE 2.0 (Lowe & Eddy Citation1997) were used to annotate this mitochondrial genome.
The mitochondrial genome of C. trimaculatus is 16,581bp in length, including 2 ribosomal RNA genes (rRNA), 22 transfer RNA genes (tRNA) and 13 protein-coding genes (PCGs). Its GC content is 46.00% (27.35% A, 26.64%T, 30.05% C, 15.97% G), similar to C. virginalis (45.74%) from the same family, Cichlidae. The lengths of 22 tRNA genes range from 67 bp (tRNACys and tRNASer) to 74 bp (tRNALeu and tRNALys), whereas 16S rRNA is 1,674bp and 12S rRNA is 940bp. All PCGs in C. trimaculatus are started with ATG and stopped with TAN or AGA, except for COX1 started with GTG.
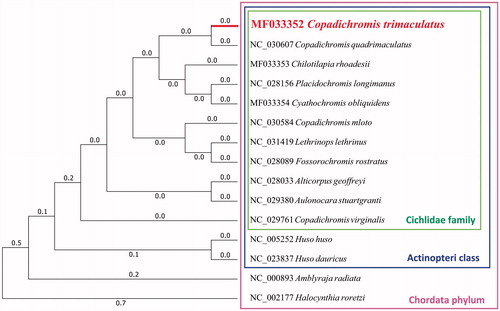
Furthermore, we used MEGA7 (V7.0.25) (Kumar et al. Citation2016) to construct the phylogenetic tree on the complete mitochondrial genomes of C. trimaculatus and other 14 related species ().
Acknowledgements
The authors would like to thank Wellcome Trust Sanger Institute (SC) for generating and uploading the raw data of the C. trimaculatus used in this study. This work was sponsored by Qing Lan project in Jiangsu Province.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33:1870–1874.
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 25:1966–1967.
- Lowe TM, Eddy S. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.