Abstract
The lesser aspen webworm moth, Meroptera pravella, is a small pyralid that uses quaking aspen (Populus tremuloides) and related tree species as larval hosts. Whole-genome Illumina sequencing allowed the assembly of a complete circular mitochondrial genome of 15,260 bp consisting of 80.7% AT nucleotides, 22 tRNAs, 13 protein-coding genes, 2 rRNAs and a control region. Mitogenome structure maintains complete synteny with other sequenced pyralid mitogenomes. Parsimony and maximum-likelihood phylogenetic reconstruction places M. pravella within monophyletic subfamily Phycitinae and monophyletic family Pyralidae. The Pyralidae, with monophyletic sister family Crambidae, constitute monophyletic superfamily Pyraloidea, which is consistent with conventional taxonomy.
The Living Prairie Mitogenomics Consortium seeks to accumulate arthropod mitochondrial genomes from a single location to produce a reference library for improved DNA-based species identification and phylogenetics (McCullagh Citation2016). Mitochondrial genome sequences were assembled and annotated by undergraduates in a course of inquiry-based learning exercise (Marcus et al. Citation2010). Students analyzing the data successfully (which were further curated by the instructor) belong to our consortium.
The Living Prairie Museum (LPM) consists of 12.9 hectares of relict unploughed prairie maintained by periodic controlled burns located in Winnipeg, Manitoba, Canada (GPS 49.889607 N, −97.270487 W). Over 160 native plant species occur at LPM, supporting a rich arthropod fauna. Arthropods were sampled weekly during the 2015 growing season.
On 17–18 July 2015, a USDA blacklight trap (Winter Citation2000) was deployed to collect night-flying insects. One adult specimen of the lesser aspen webworm moth Meroptera pravella (Pyralidae, project specimen number 2015.07.17.012) was trapped. This specimen likely originated from a nearby grove of quaking aspen (Populus tremuloides) larval host plant located at LPM. The specimen was pinned, spread and deposited in the collection of the Wallis Roughley Museum of Entomology at the University of Manitoba (voucher JBWM0363025).
DNA was prepared (McCullagh & Marcus Citation2015) and sequenced by Illumina MiSeq (San Diego, CA) (Peters & Marcus Citation2017). Overall, 2,958,411 paired reads (total 1.7 Gb) were assembled in Geneious 10.1.2 to a Plodia interpunctella (Pyralidae) reference mitogenome (KT207942.1) to reconstruct a complete mitogenome sequence for M. pravella (GenBank MF073207). Annotation was performed with reference to P. interpunctella and Junonia lemonias (Nymphalidae, KP941756) mitogenomes (McCullagh & Marcus Citation2015). The complete M. pravella nuclear rRNA repeat (GenBank MF073208) was also assembled and annotated with respect to the Attacus ricini rRNA repeat (Saturniidae, AF463459)
The circular mitogenome of M. pravella consists of 15,260 bp with nucleotide composition of 39.5% A, 11.6% C, 7.7% G and 41.0% T. Meroptera pravella maintains complete synteny with other mitogenomes from superfamily Pyraloidea and other Ditrysian Lepidoptera (Cao et al. Citation2012). Meroptera pravella COI has an aberrant start codon (CGA) that is typical of insects (Peters & Marcus Citation2016). Four mitochondrial protein-coding genes (NAD2, COI, COII and NAD5) have aberrant single-nucleotide (T) stop codons. As in many other arthropods, all M. pravella tRNAs have standard cloverleaf secondary structures except for trnS (AGN) which has the dihydrouridine arm replaced by a loop (McCullagh & Marcus Citation2015). The rRNAs (775 bp 12S and 1382 bp 16s) are composed of 84.7% AT while the putative control region (278 bp) is 96.0% AT.
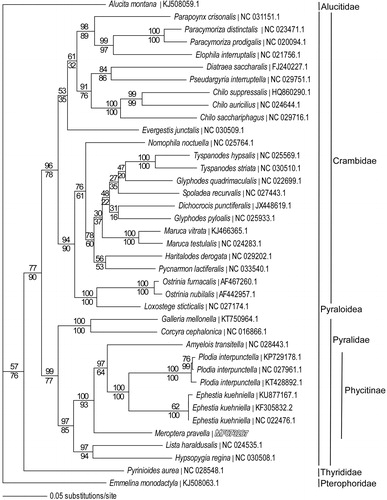
We reconstructed a phylogeny using mitogenomes from M. pravella, 7 other Pyralid moth species, 24 Crambidae species and representatives from the related families Thyrididae, Alucitidae and Pterophoridae. Sequences were aligned in CLUSTAL Omega (Sievers et al. Citation2011) and analyzed by parsimony and maximum likelihood in PAUP* 4.0b8/4.0d78 (Swofford Citation2002) (). Phylogenetic analysis places M. pravella as the basal lineage within subfamily Phycitinae of family Pyralidae and supports the conventional sister taxon relationship between monophyletic families Pyralidae and Crambidae in superfamily Pyraloidea (Munroe & Solis Citation1998).
Figure 1. Maximum-likelihood phylogeny (GTR + I + G model, I = 0.2790, G = 0.4760, likelihood score 190,748.88) of Meroptera pravella and related species in families Pyralidae, Crambidae, Thyrididae, Alucitidae and Pterophoridae based on one million random addition heuristic search replicates (with tree bisection and reconnection) of aligned complete mitochondrial genomes. One million maximum parsimony heuristic search replicates produced a nearly identical tree topology for family Pyralidae (parsimony score 41,119 steps), but with a monophyletic Glyphodes and with Evergestis as sister to the Diatraea-Pseudargyria-Chilo clade in the Crambidae. Numbers above each node are maximum-likelihood bootstrap values and numbers below each node are maximum parsimony bootstrap values (each from one million random fast addition search replicates).
Acknowledgements
We thank Sarah Semmler and Kyle Lucyk for permitting and encouraging our work at the Living Prairie Museum. We thank Aleksandar Ilik and Debbie Tsuyuki (Children’s Hospital Research Institute of Manitoba Next Generation Sequencing Platform) for assistance with library preparation and sequencing. This work received support from the Teaching and Learning Enhancement Fund of the University of Manitoba Centre for the Advancement of Teaching and Learning and NSERC under Grants RGPIN386337-2011 and RGPIN-2016-06012.
Disclosure statement
The authors report no conflicts of interest and are solely responsible for this paper.
Additional information
Funding
References
- Cao YQ, Ma C, Chen JY, Yang DR. 2012. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in lepidoptera. BMC Genom. 13:276.
- Marcus JM, Hughes TM, McElroy DM, Wyatt RE. 2010. Engaging first year undergraduates in hands-on research experiences: the Upper Green River Barcode of Life Project. J Coll Sci Teach. 39:39–45.
- McCullagh BS, Marcus JM. 2015. The complete mitochondrional genome of Lemon Pansy, Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae). J Asia-Pacific Ent. 18:749–755.
- McCullagh BS. 2016. Sequence evolution among divergent mitochondrial haplotypes within species of Junonia butterflies. M.Sc. Thesis. Winnipeg (MB) Canada: Department of Biological Sciences, University of Manitoba.
- Munroe EG, Solis MA. 1998. The pyraloidea. In: Kristensen NP, editor. Lepidoptera, moths and butterflies volume 1: evolution, systematics, and biogeography. Berlin (Germany): Walter de Gruyter; p. 233–256.
- Peters MJ, Marcus JM. 2016. The complete mitochondrial genome of the Bermuda buckeye butterfly Junonia coenia bergi (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B. 1:739–741.
- Peters MJ, Marcus JM. 2017. Taxonomy as a hypothesis: testing the status of the Bermuda buckeye butterfly Junonia coenia bergi (Lepidoptera: Nymphalidae). Syst Ent. 42:288–300.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.
- Swofford DL. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates; Sunderland, MA.
- Winter WD. 2000. Basic techniques for observing and studying moths and butterflies. Vol. 5. Los Angeles (CA): The Lepidopterists' Society.
