Abstract
The complete mitochondrial genome of Anas crecca is 16,596 bp in length. It was predicted to contain 13 PCGs, 22 tRNA genes, and 2 rRNA genes, and a putative control region. All of the PCGs initiate with ATG, except for MT-CO1, MT-CO2, and MT-ND5, which began with GTG. Four types of termination codons were identified. Phylogenetic analysis shows that A. crecca clustered with Anas acuta, and the monophyly of the genera Anser, Cygnus, Aythya, and Anas was strongly supported. Moreover, our results also support a sister-group relationship between Anas and Aythya.
Anatidae is the largest family in Anseriformes among the three families (Anhimidae, Anseranatidae, and Anatidae). About 173 species in 49 genera are found in this family (Gill and Donsker Citation2017). Anas is a relatively large genus of the family Anatidae (Clements et al. Citation2016). In this study, an adult Anas crecca was collected from Wuhu (31.33°N, 118.38°E) of southeastern China, and the specimen was stored in the herbarium of Anhui Normal University. The total genomic DNA of the A. crecca (code Kan-A0147) was extracted from the muscle tissue with standard phenol/chloroform methods. Fourteen PCR and sequencing primers specific to A. crecca were designed, and two long overlapping fragments were amplified to avoid the possibility of obtaining nuclear copies of mitochondrial genes (Zhang et al. Citation2012).
We obtained the complete mitochondrial genome of A. crecca (GenBank Accession No. KC771255) in this study. We described 16,596 bp of A. crecca mitochondrial genome DNA, including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 putative control region. All tRNA genes can fold into a typical cloverleaf second structure. All of the PCGs initiate with ATG, except for MT-CO1, MT-CO2, and MT-ND5, which began with GTG. Four types of termination codons were identified, canonical TAA and TAG termination codons are found in eight PCGs, while AGG for MT-CO1 and MT-ND1, the remaining genes (MT-ND2, MT-CO3, and MT-ND4) terminate with an incomplete stop codon (T––). We can detect a single control region (D-loop) between tRNAGlu and tRNAPhe. The D-loop region is 1044 bp in length. The overall base composition of the mitochondrial genome is as follows: A (28.99%), T (22.37%), G (15.96%), and C (32.67%), with the A + T content of 51.37%. Nucleotide composition was calculated by MEGA6 (V6.06) (Tamura et al. Citation2013). The MT-ND6 gene of A. crecca mitogenome has strong skews of T versus A (−0.57) (Kan et al. Citation2010), and a strong skew of C versus G (0.53). The other 12 PCGs of the A. crecca mitochondrial genome have a slight skew of A versus T (0–0.32), and a strong skew of C versus G (GC skew= −0.65 to −0.31).
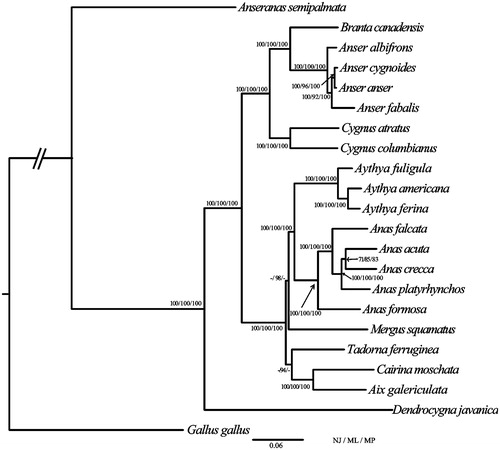
The phylogenetic position of A. crecca was estimated from a concatenated dataset based on 13 PCGs (Kan et al. Citation2010). Maximum likelihood (ML) analyses were performed with raxmlGUI (V1.3.1) (Silvestro and Michalak Citation2012; Jiang et al. Citation2015). Neighbour-Joining (NJ) analyses were conducted in MEGA6 (V6.06), and Maximum parsimony (MP) analyses were calculated using PAUP* (V4.0 b 10) (Swofford Citation2003). The complete mitochondrial DNA sequences have been used successfully to estimate phylogenetic relationships among Anseriformes. Our results show that A. crecca clustered with A. acuta, the bootstrap value was 85% in ML analyses. Our results also indicate that the monophyly of Anser, Cygnus, Aythya, and Anas was strongly supported, respectively. Moreover, our results found a sister-group relationship between Anas and Aythya, and Anseranas was basal to the balance of the Anseriformes. This study will contribute to the phylogenetic analyses in Anseriformes ().
Figure 1. Phylogenetic tree of the relationships among Anseriformes based on the nucleotide dataset of the 13 PCGs. Gallus gallus served as outgroup. The NJ analyses were conducted using 1000 bootstrap values in MEGA v 6.06, ML analyses were implemented in ML + rapid bootstrap for 100 replicates under GTRGAMMA, and MP analyses were performed with 1000 bootstrap values. Number of above each node indicates the NJ, ML, and MP bootstrap support values, respectively. Branch lengths and topologies came from the ML analysis. All 22 species' accession numbers are listed as below: Gallus gallus KM096864, Anseranas semipalmata NC_005933, Dendrocygna javanica NC_012844, Aix galericulata NC_023969, Cairina moschata NC_010965, Tadorna ferruginea NC_024640, Mergus squamatus NC_016723, Anas formosa NC_015482, Anas platyrhynchos NC_009684, Anas acuta NC_024631, Anas falcata NC_023352, Aythya ferina NC_024602, Aythya americana NC_000877, Aythya fuligula NC_024595, Cygnus columbianus NC_007691, Cygnus atratus NC_012843, Anser fabalis NC_016922, Anser anser NC_011196, Anser cygnoides NC_023832, Anser albifrons NC_004539, Branta canadensis NC_007011, and Anas crecca KC771255.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Clements J, Schulenberg T, Iliff M, Roberson D, Fredericks T, Sullivan B, Wood C. 2016. The eBird/Clements checklist of birds of the world: v2016. Available from: http://www.birds.cornell.edu/clementschecklist/download/.
- Gill F, Donsker D. 2017. IOC World Bird List (v 7.2). doi: 10.14344/IOC.ML.7.2.
- Jiang L, Chen J, Wang P, Ren Q, Yuan J, Qian C, Hua X, Guo Z, Zhang L, Yang J. 2015. The mitochondrial genomes of Aquila fasciata and Buteo lagopus (Aves, Accipitriformes): sequence, structure and phylogenetic analyses. PLoS One. 10:e0136297.
- Kan X, Li X, Lei Z, Wang M, Chen L, Gao H, Yang Z. 2010. Complete mitochondrial genome of Cabot’s tragopan, Tragopan caboti (Galliformes: Phasianidae). Genet Mol Res. 9:1204–1216.
- Kan X, Yang J, Li X, Chen L, Lei Z, Wang M, Qian C, Gao H, Yang Z. 2010. Phylogeny of major lineages of galliform birds (Aves: Galliformes) based on complete mitochondrial genomes. Genet Mol Res. 9:1625–1633.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Organ Divers Evol. 12:335–337.
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zhang L, Wang L, Gowda V, Wang M, Li X, Kan X. 2012. The mitochondrial genome of the Cinnamon Bittern, Ixobrychus cinnamomeus (Pelecaniformes: Ardeidae): sequence, structure and phylogenetic analysis. Mol Biol Rep. 39:8315–8326.
