Abstract
The complete mitochondrial genome of Thermophis baileyi was sequenced using the next-generation sequencing (NGS) in the present study. The total length of the mitogenome was 17,355 bp, which was composed of 13 protein-coding genes, 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes (12s and 16s rRNA), and 2 control regions (CR I and CR II). The base composition was 32.4% for A, 23.8% for T, 30.2% for C, and 13.6% for G. Coding genes of each protein in the mtDNA had the same start and stop codons among three Thermophis species.
Genus Thermophis contains only three species (Thermophis zhaoermii, Thermophis shangrila, Thermophis baileyi), which is distributed in a few sites at high altitudes (over 3500 m) (Huang et al. Citation2009; He et al. Citation2010; Alex et al. Citation2016). Thermophis baileyi is endemic to the Tibetan Plateau and was firstly described by Wall (1907) based on characters (Zhao Citation2008; Hofmann Citation2012). In this study, we determined the complete mitochondrial genome of T. baileyi using the next-generation sequencing (NGS).
Sample was collected from the Yangbajing, Xizang Privonce, China. Specimen was stored in the CIB herpetological museum, Chengdu Institute of Biology with the number of CIBLJT20150701. The genomic DNA isolated from the sample was sequenced using NGS (Hahn et al. Citation2013). We predicted and analyzed the mitogenomic structure using MITOS web server (Bernt et al. Citation2013). RefSeqs which were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/genome/organelle/) was used to correct the mitogenomic structure manually. The tRNA genes were scanned by tRNAscan-SE (Lowe and Eddy Citation1997). MEGA5 was used to calculate the base composition and to align the whole-genome sequence data (Tamura et al. Citation2001). Maximum likelihood (ML) method was used to reconstruct a phylogenetic tree in the present study by using MEGA5 (Tamura et al. Citation2001).
The mitogenome of T. baileyi (GenBank MF326642) was 17,355 bp in length, with a base composition of 32.4% for A, 23.8% for T, 30.2% for C, and 13.6% for G. The mitogenome sequenced by us contained 13 protein-coding genes, 2 rRNA genes (12s and 16s rRNA), 22 tRNA genes, and 2 control regions (CR I and CR II). The ND 6 gene encoded on the light-strand, with the remaining protein coding genes encoding on the heavy strand. Compared with the other two hot-spring snakes (Thermophis zhaoermii NC012816; Thermophis shangrila MF066951), each protein-coding gene in the mtDNA had the same start and stop codon among three Thermophis species (He et al. Citation2010; Wu et al. Citation2017). The length of CRI was 1129 bp, ranging from 16,226 to 17,355 bp and CRII was 1027 bp, ranging from 3629 to 4655 bp. The length of 22 tRNA genes ranged from 57 bp to 73 bp.
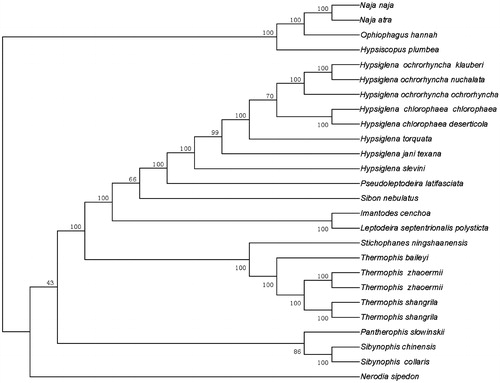
A ML tree based on the total mitochondrial genome sequences from 26 species of snakes supported that Thermophis baileyi, Thermophis zhaoermii and Thermophis shangrila form a molophylitic clade (). The evolutionary relationships of these species were consistent with previously reported results (Alex et al. Citation2016). The mitogenome characterization of the T. baileyi was helpful to understand the evolution of Colubridae.
Figure 1. Molecular phylogeny of Thermophis baileyi and other related species in ferns based on complete mitochondrial genome. The sequences accession number for tree re-construction is listed as follows: Thermophis baileyi (MF326642); Hypsiscopus plumbea (DQ343650); Nerodia sipedon (JF964960); Pantherophis slowinskii (DQ523162); Sibynophis chinensis (NC022430); Sibynophis collaris (NC016424); Thermophis zhaoermii (GQ166168); Thermophis zhaoermii (NC012816); Thermophis shangrila (MF066951); Thermophis shangrila (KU174488); Pseudoleptodeira latifasciata (EU728579); Hypsiglena chlorophaea chlorophaea (EU728593); Hypsiglena chlorophaea deserticola (EU728587); Hypsiglena jani texana (EU728592); Hypsiglena ochrorhyncha klauberi (EU728589); Hypsiglena ochrorhyncha nuchalata (EU728581); Hypsiglena ochrorhyncha ochrorhyncha (EU728578); Hypsiglena slevini (NC013987); Hypsiglena torquata (EU728591); Imantodes cenchoa (EU728586); Sibon nebulatus (EU728583); Leptodeira septentrionalis polysticta (EU728590); Stichophanes ningshaanensis (NC026083); Naja atra (EU913475); Ophiophagus hannah (NC011394); Naja naja (DQ343648).
Acknowledgements
We thank Ke Jiang and Wei Wu for assistance in sample collection and the phylogeny analysis.
Disclosure statement
The author has declared that no conflict of interest exists. The author is responsible for the content and writing of the paper.
Additional information
Funding
Reference
- Alex F, Mckelvy AD, Lee GL, Bell CD, Lailvaux SP. 2016. A species-level phylogeny of extant snakes with description of a new colubrid subfamily and genus. PLoS One. 11:1–31.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- He M, Feng JC, Zhao EM. 2010. The complete mitochondrial genome of the Sichuan hot-spring keel-back (Thermophis zhaoermii; Serpentes: Colubridae) and a mitogenomic phylogeny of the snakes. Mitochondr DNA. 21:8–18.
- Hofmann S. 2012. Population genetic structure and geographic differentiation in the hot spring snake Thermophis baileyi (Serpentes, Colubridae): indications for glacial refuges in southern-central Tibet. Mol Phylogenet Evol. 63:396–406.
- Huang S, Liu SY, Guo P, Zhang YP, Zhao EM. 2009. What are the closest relatives of the hot-spring snakes (Colubridae, Thermophis), the relict species endemic to the Tibetan Plateau? Mol Phylogenet Evol. 51:438–446.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2001. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Wu W, Jiang DC, Sun FH. 2017. Next-generation sequencing yields the complete mitochondrial genome of the Shangrila hot-spring snakes (Thermophis shangrila; Reptilia: Colubridae). Mitochondr DNA PART B. Resources. 2:327–328.
- Zhao EM. 2008. The World-specific snakes at Qinghai - Tibet Plateau, hot-spring snakes. J Cun. 17:5–9.
