Abstract
We have determined the mitochondrial genome of Reticulitermes kanmonensis Takematsu, 1999. The total length of the R. kanmonensis is 16,484 bp with 66.1% A + T content. It consists of 13 PCGs, 22 tRNA, 2 rRNA genes and an A + T–rich control region. All the protein-coding genes used ATN as start codon. But the stop codons were TAA, TAG, and an incomplete termination codon (T) abutting an adjacent tRNA gene. The A + T–rich control region was 1680 bp in length with 70.4% A + T content.
Reticulitermes kanmonensis Takematsu (Citation1999) is an economically important termite species in Japan. It has been responsible for damaging traditional wooden houses or cultural properties in Japan (Takematsu Citation1999). In Korea, the distribution of this species was firstly reported in 2014 (Lee et al. Citation2015). Although there are some taxonomic studies about this termite (Takematsu Citation1999; Lee et al. Citation2015), there is no available information for the mitogenome of this species.
The present study reported the complete mitogenome sequences of R. kanmonensis. Specimens were collected from Wanju-gun, Jeollabuk-do, Korea, in April 2014. Voucher specimens (Coll#140428WH08) were deposited at the Insect Collection, Gyeongsang National University, Korea. Total genomic DNA was extracted from the soldier of R. kanmonensis. The mitogenome was determined by sequencing with Illumina HiSeq4000, Macrogen, Inc., Korea. Raw sequences were filtered by Trimmomatic and aligned with mitogenome of R. speratus kyushuensis (Lee et al. Citation2017). The mitogenome of R. kanmonensis is 16,484 bp in size and its gene content and organization were identical with other Reticulitermes species, presenting 13 PCGs, 22 tRNA genes, 2 rRNA genes, and an A + T–rich control region. The N chain coded 14 gene, including 8 tRNA genes (tRNAGln, tRNACys, tRNATyr, tRNAPhe, tRNAHis, tRNAPro, tRNALeu(CUN), and tRNAVal), four PCGs (ND5, ND4, ND4L, and ND1), and two rRNA genes (lrRNA and srRNA). The rest 23 genes were coded by the J chain.
The overall sequences in the mitogenome of R. kanmonensis were A + T–biased (the G + C content was 33.9%), as commonly observed in insect mitogenomes. The PCGs region and A + T–rich region accounted for 64.5% AT and 70.4% AT, respectively. R. kanmonensis had the typical 22 tRNA genes throughout the entire mitogenome. All of them, except that tRNASer(AGN) lacked the dihydrouridine (DHU) stem, showed the typical clover-leaf secondary structure. Thirteen open reading frames of R. kanmonensis protein-coding sequences had typical ATN initiation codon. Nine genes had a complete TAA termination codon and two genes (ND1 and ND2) had a complete TAG termination codon. While two genes (COX2 and ND5) showed an incomplete termination codon (T) abutting an adjacent tRNA gene, which is commonly observed in metazoan animals.
Mitogenome of R. kanmonensis also included intergenic spacers and overlapping regions in common with other termite species. The intergenic spacer sequences were spread on 18 regions ranging in size from 1 to 19 bp, and the overlapping sequences varied from 1 to 19 bp in 6 areas. The A + T–rich control region of R. kanmonensis located between srRNA and tRNAIle with 1680 bp long and 70.4% A + T content.
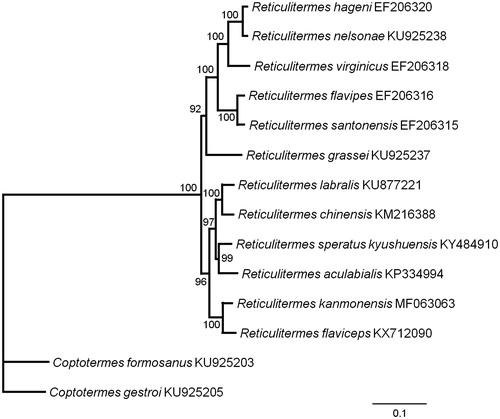
Nucleotide sequences of 13 PCGs from 13 closely related species were analyzed to investigate phylogenetic relationships with R. kanmonensis. Phylogenetic analysis was performed using maximum likelihood method with 1000 bootstrap replications with PhyML 3.0 (Guindon et al. Citation2010). The phylogenetic tree revealed that Reticulitermes was a sister group to Coptotermes, and R. kanmonensis and Reticulitermes flaviceps form one clade (). Our study of R. kanmonensis will be a useful research for understanding the classification and status in Rhinotermitidae.
Figure 1. Maximum likelihood estimation of the phylogenetic relationships of two genera Reticulitermes and Coptotermes based on the nucleotide sequence of 13 PCGs in the mitochondrial genome. The numbers beside the nodes are percentages of 1000 bootstrap values. The Coptotermes formosanus and Coptotermes gestroi were used as an outgroup. Alphanumeric terms indicate the GenBank accession numbers.
Nucleotide sequence accession number
The complete mitochondrial genome sequence of R. kanmonensis has been assigned GenBank accession number MF063063.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Guindon S, Dufayard J–F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Lee W, Choi D–S, Ji J–Y, Kim N, Han JM, Park S–H, Lee S, Seo MS, Hwang W–J, Forschler BT, et al. 2015. A new record of Reticulitermes kanmonensis Takematsu,1999 (Isoptera: Rhinotermitidae) from Korea. J Asia Pac Entomol. 18:351–359.
- Lee W, Han T, Lee J-H, Hong K-J, Park J. 2017. The complete mitochondrial genome of the subterranean termite, Reticulitermes speratus kyushuensis Morimoto, 1968 (Isoptera: Rhinotermitidae). Mitochondrial DNA Part B. 2:178–179.
- Takematsu Y. 1999. The genus Reticulitermes (Isoptera: Rhinotermitidae) in Japan, with description of a new species. Entomol Sci. 2:231–243.
