Abstract
The mitogenome of the Asian badger Meles leucurus from Korea is a circular molecule of 16,529 bp, consisting of a control region and a conserved set of 37 genes containing 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes (12S rRNA and 16S rRNA). The mitogenome of M. leucurus is AT-biased, with a nucleotide composition of 33.1% A, 27.9% T, 25.4% C, and 13.5% G. The phylogenetic analysis revealed that the badger M. leucurus from Korea is well grouped with that from China, forming a sister clade to M. meles from Japan.
The Asian badger Meles leucurus, also known as the sand badger, is found in Mongolia, China, Kazakhstan, the Korean Peninsula, and Russia (Abramov Citation2016). We sequenced and characterized the complete mitogenome of the Asian badger M. leucurus (Mustelidae) from Korea. Genomic DNA was extracted from a road-killed individual around agroecosystem in Odaesan National Park (N37 45 26.4, E128 36 39.8). The voucher specimen (MUMELE-1) was deposited in the National Park Research Institute, Korea National Park Service. Genomic DNA extraction, PCR, and gene annotation were conducted according to the previous studies (Yoon et al. Citation2013; Jeon and Park Citation2015; Rahman et al. Citation2016). Previously published mitogenomes of the Eurasian M. meles (AM711900) (Arnason et al. Citation2007) and Japanese M. anakuma (AB291075) (Yonezawa et al. Citation2007) were used as references for gene annotation and primer design for PCR amplification of the Korean M. leucurus. Phylogenetic tree was constructed using maximum likelihood (ML) procedures implemented in MEGA6 (Tamura et al. Citation2013).
The complete mitogenome (MF497304) of M. leucurus is 16,529 bp total length, which consists of a CR and a conserved set of 37 vertebrate mitochondrial genes including 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes (12S rRNA and 16S rRNA). The order and orientation of these genes are identical to those other mammalian species (Kim and Park Citation2012; Yoon et al. Citation2013; Nam et al. Citation2015). The mitogenome of M. leucurus is AT-biased, with a nucleotide composition of 33.1% A, 27.9% T, 25.4% C, and 13.5% G.
Total length of 13 PCGs consists of 11,382 bp, with the exclusion of stop codons, which encode 3794 amino acids. The start codon ATG is used in all the other PCGs except for Nd2 (ATT), Nd3 (ATA), and Nd5 (ATC). The stop codon TAA is used for termination of seven PCGs (Cox1, Cox2, Atp8, Atp6, Nd4L, Nd5, and Nd6), while AGA and TAG occur once in Cytb and Nd3, respectively. Incomplete stop codon T– is found in Nd1, Nd2, Cox3, and Nd4. Ribosomal RNA genes (12srRNA and 16srRNA) are located between tRNAPhe and tRNALeu(CUN) and separated by tRNAVal. Lengths of 12S rRNA and 16S rRNA gene were 960 bp and 1573 bp long, respectively. Length of 22 tRNA genes for transferring 20 amino acids ranges from 62 bp (tRNASer (AGY)) to 75 bp (tRNALeu (UUR)). Lengths of the two-non coding genes are 1088 bp (control region) and 36 bp (OL) long, respectively.
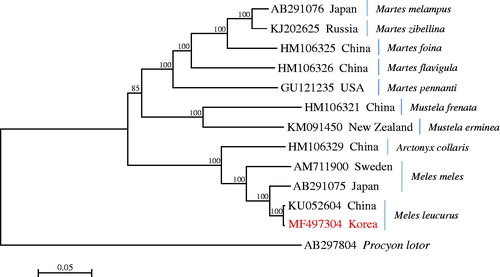
The phylogenetic analysis revealed that the badger M. leucurus from Korea is well grouped with that from China, forming a sister clade to M. meles from Japan. The clade of the genus Meles was strongly supported with high bootstrap values and formed a sister clade to the hog badger Arctonyx collaris ().
Figure 1. The phylogenetic relationship of the Asian badger Meles leucurus and its allied species inferred from maximum-likelihood analysis based on mitogenome sequences. The Korean badger M. leucurus used in this study was indicated with the mitogenome accession no. MF497304. The ML tree was generated using the GTR + G+I model, and the robustness of the tree was tested with 1000 bootstrap. The numbers on the branches indicate bootstrap values.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Abramov AV. 2016. Meles leucurus. The IUCN Red List of Threatened Species 2016: e.T136385A45221149 [accessed 2017 July 19].
- Arnason A, Andersen T, Holme I, Engebretsen L, Bahr R. 2007. Prevention of hamstring strains in elite soccer: an intervention study. Scand J Med Sci Sports. 18:40–48.
- Jeon MG, Park YC. 2015. The complete mitogenome of the wood-feeding cockroach Cryptocercus kyebangensis (Blattodea: Cryptocercidae) and phylogenetic relations among cockroach families. Anim Cells Syst. 19:432–438.
- Kim HR, Park YC. 2012. The complete mitochondrial genome of the striped field mouse, Apodemus agrarius (Rodentia, Murinae) from Korea. Mitochondr DNA. 23:145–147.
- Nam TW, Kim HR, Cho JY, Park YC. 2015. Complete mitochondrial genome of a large-footed bat, Myotis macrodactylus (Vespertilionidae). Mitochondrial DNA. 26:661–662.
- Rahman MM, Yoon KB, Kim JY, Hussin MZ, Park YC. 2016. Complete mitochondrial genome sequence of the Indian pipistrelle Pipistrellus coromandra (Vespertilioninae). Anim Cells Syst. 20:86–94.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Yonezawa D, Sekiguchi F, Miyamoto M, Taniguchi E, Honjo M, Masuko T, Nishikawa H, Kawabata A. 2007. A protective role of hydrogen sulfide against oxidative stress in rat gastric mucosal epithelium. Toxicology. 241:11–18.
- Yoon KB, Cho JY, Park YC. 2013. Complete mitochondrial genome of the Korean ikonnikov’s bat Myotis ikonnikovi (Chiroptera: Vespertilionidae). Mitochondr DNA. 26:274–275.
