Abstract
Giant-Skipper butterflies from the genus Agathymus (family Hesperiidae) are unusual as their caterpillars feed inside Agave leaves. Relationships among Agathymus taxa and their names (i.e. if they are species, subspecies, or synonyms) are poorly understood due to phenotypic similarity. DNA sequences are promising to clarify the taxonomic questions, but it is challenging to sequence name-bearing types that are usually old specimens with poorly preserved DNA. Using next generation sequencing, we assembled mitochondrial genomes of four Agathymus mariae group holotype specimens collected more than 55 years ago and housed pinned and dry in the American Museum of Natural History (New York, NY). We compared the holotype mitogenomes to those we obtained from fresh A. mariae specimens and the sister species Agathymus micheneri. All but A. micheneri mitogenomes were highly similar to each other (more than 99% identity), suggesting that the four names chinatiensis, lajitaensis, rindgei, and gilberti proposed by H. A. Freeman in 1964 may not refer to species-level taxa. The mitogenomes grouped eastern populations (rindgei and gilberti) together and apart from the western populations (nominal mariae, chinatiensis, and lajitaensis). Mexican A. micheneri differs by about 2.5% (about 5% in the COI barcode region) from A. mariae, and is likely to be a distinct species.
The Giant-Skippers (Hesperiidae: Megathymini) are endemics of the North American continent. They are known for their large size and unusual life habits. Adults do not feed and caterpillars live inside their foodplants: in Yucca roots and Agave leaves (Freeman Citation1969; Roever Citation1975; Scott Citation1986). Agave-feeding Agathymus is the most species-rich genus and its taxonomy is far from clear (Roever Citation1975; Scott Citation1986; Roever Citation1998). It remains to be studied which proposed names denote species, subspecies or synonyms. Agathymus mariae group includes skippers with caterpillars feeding on lechuguilla and distributed in western Texas, southern New Mexico and north-central Mexico (Freeman Citation1969). Six names exist for the populations in this group (Freeman Citation1969; Mielke Citation2005; Pelham Citation2008). In addition to nominal A. mariae (type locality: Texas, El Paso) (Barnes and Benjamin Citation1924) and Mexican A. micheneri (type locality: Mexico, Coahuila) (Stallings et al. Citation1961), four new taxa were proposed in 1964 as full species by H. A. Freeman (Freeman Citation1964). Agathymus rindgei and A. gilberti are sympatric with the same type locality (Texas, Kinney County, 14 mi north of Brackettville) and represent eastern populations of the mariae group. Agathymus rindgei and A. gilberti were suggested to have different number of chromosomes that correlated with the darker appearance and smaller yellow-orange spots of A. gilberti. Agathymus chinatiensis and lajitaensis, both from Presidio County in Texas, together with the nominal A. mariae constitute western populations and have more prominent pale spots on the hindwing below (Freeman Citation1969). As all mariae group taxa are morphologically similar, subsequent authors suggested that most, if not all, existing names correspond to either subspecies or synonyms (Roever Citation1975; Scott Citation1986; Pelham Citation2008). To further understand the relationships between these taxa, it would be instructive to investigate DNA differences between them.
Since the ultimate taxonomic study should be based on the analysis of name-bearing specimens, we sequenced, assembled and annotated the complete mitogenomes of the holotypes of Freeman's A. mariae group names. All four specimens have been housed pinned and dry in the American Museum of Natural History collection (New York, NY). Since the genitalia of all holotypes have been dissected, a small amount of tissue was sampled from the end of the abdomen through the cut introduced at the time of dissection. Methods for genomic DNA extraction, library construction, next-generation sequencing, and computational procedures have been reported by us previously (Shen et al. Citation2015; Cong et al. Citation2016a, Citation2016b; Shen et al. Citation2016; Zhang et al. Citation2017a, Citation2017b, Citation2017c). The mitogenome of Agathymus mariae mariae was used as a reference to search for (‘bait’) similar sequence reads using BWA (Li and Durbin Citation2009). The mitogenomes were assembled from these reads de novo using Platanus (Kajitani et al. Citation2014) followed by a manual gap-closing procedure. Due to possibly variable number of direct repeats that we previously found in the D-loop region (Zhang et al. Citation2017b), exact sequence of this region is uncertain in all Megathymini mitogenomes.
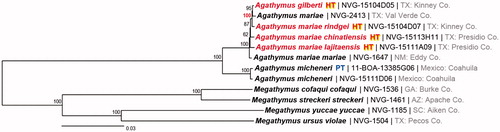
To assess the quality of the genomes of the holotypes collected 55 years ago, we sequenced a mitogenome of a fresh A. mariae group specimen from the eastern part of the range (voucher NVG-2413 from Texas: Val Verde County) and used the sequence we obtained previously (Zhang et al. Citation2017b) from a fresh A. mariae group specimen from the western part of the range. Finally, mitogenomes of two Agathymus micheneri specimens (one is a paratype) from Mexico: Coahuila, were sequenced using the same methods. Phylogenetic tree constructed from the mitogenomes reveals that A. micheneri is the most distant taxon from the rest, showing about 2.5% difference in mitogenome regions excluding D-loop and about 5% difference in the COI barcode region (). Although originally proposed as a subspecies of A. mariae, A. micheneri is mostly treated as a full species in recent literature. Differences in its mitogenome support the status of A. micheneri as a species. Interestingly, the differences among mitogenomes of all other taxa are minimal: less than 0.6% in regions excluding D-loop and 0.7% in the COI barcode region, which is consistent with their possible conspecificity. Specimens group by locality rather than by age (i.e. fresh specimens are not clustered together in ), which supports sufficient quality of DNA sequences obtained from half-a-century-old specimens. Notably, specimens from the eastern (Kinney & Val Verde Counties) populations group together with 100 bootstrap support. Sympatric A. gilberti and rindgei did not reveal prominent differences in their mitogenomes, and divergence between them (0.44%) is not larger than that among named A. mariae subspecies (0.42–0.59%). As sympatric taxa cannot be subspecies of the same species, A. gilberti and rindgei are either synonymous, or mitochondrial introgression complicates the assessment, and the analysis of nuclear genes is necessary to fully resolve this taxonomic puzzle.
Figure 1. Maximum likelihood tree of mitogenomes of 12 Megathymini specimens. Specimen numbers are shown after the names followed by a general locality. Numbers by the nodes show bootstrap support values. GenBank accessions for sequences and data for Agathymus specimens are: Agathymus gilberti holotype, MF684854, voucher NVG-15104D05, female, USA: Texas, Kinney County, 14 mi N of Brackettville, elevation 1500′, 22 October 1961; Agathymus mariae chinatiensis holotype, MF684857, voucher NVG-15113H11, female, USA: Texas, Presidio County, 2.7 mi S of Shafter, elevation 4000′, 5 October 1960; Agathymus mariae lajitaensis holotype, MF684856, voucher NVG-15111A09, female, USA: Texas, Presidio County, 38 mi SE of Presidio, elevation 2650′, 2 October 1961; Agathymus mariae mariae KY630504, voucher NVG-1647, female, USA: New Mexico, Eddy County, 22-Sep-2013; Agathymus mariae rindgei holotype, MF684855, voucher NVG-15104D07, female, USA: Texas, Kinney County, 14 mi N of Brackettville, elevation 1508′, 23 October 1961; Agathymus mariae MF684859, voucher NVG-2413, female, USA: Texas, Val Verde County, SH163 S of Juno, 26 November 2013; Agathymus micheneri paratype, MF684860, voucher 11-BOA-13385G06, male, Mexico: Coahuila, 15–20 miles south of Allende, on highway 57, Km. 8, elevation 1300′, 2 October 1957; Agathymus micheneri MF684858, voucher NVG-15111D06, female, Mexico: Coahuila, 12 mi S Allende, 2 November 1964; Megathymus cofaqui cofaqui KY630503; Megathymus streckeri streckeri KY630501; Megathymus ursus violae KY630502; Megathymus yuccae yuccae KY630500.
Acknowledgements
We are indebted to David Grimaldi and Courtney Richenbacher (American Museum of Natural History, New York, NY) and Robert K. Robbins, John M. Burns, and Brian Harris (National Museum of Natural History, Smithsonian Institution, Washington, DC) for granting access to the collections under their care, help, and stimulating discussions.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Barnes W, Benjamin FH. 1924. Notes and new species. Contrib Nat Hist Lepidoptera N Am. 5:99–199.
- Cong Q, Shen J, Borek D, Robbins RK, Otwinowski Z, Grishin NV. 2016a. Speciation in cloudless sulphurs gleaned from complete genomes. Genome Biol Evol. 8:915–931.
- Cong, Q, Shen J, Borek D, Robbins RK, Otwinowski Z, Grishin NV. 2016b. Complete genomes of Hairstreak butterflies, their speciation, and nucleo-mitochondrial incongruence. Sci Rep. 6:24863.
- Freeman HA. 1964. Four new species of Agathymus from Texas. J Lep Soc. 18:171–185.
- Freeman HA. 1969. Systematic review of the Megathymidae. J Lep Soc. 23:1–59.
- Kajitani R, Toshimoto K, Noguchi H, Toyoda A, Ogura Y, Okuno M, Yabana M, Harada M, Nagayasu E, Maruyama H, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24:1384–1395.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760.
- Mielke OHH. 2005. Catalogue of the American Hesperioidea: Hesperiidae (Lepidoptera). Brazil: Sociedade Brasileira de Zoologia. p. xiii + 1536.
- Pelham JP. 2008. Catalogue of the butterflies of the United States and Canada: with a complete bibliography of the descriptive and systematic literature. J Res Lepidoptera. 40:1–672.
- Roever K. 1975. In: Howe WH, editor. The butterflies of North America. Garden City, New York: Doubleday and Co.; p. 411–422.
- Roever K. 1998. Descriptions of new Agathymus (Lepidoptera: Megathymidae) from the southwestern United States. In: Emmel T, editor. Systematics of Western North American Butterflies. Gainesville, Florida, USA: Mariposa Press; p. 491–500.
- Scott JA. 1986. The butterflies of North America: a natural history and field guide. Stanford: Standford University Press.
- Shen J, Cong Q, Grishin NV. 2015. The complete mitochondrial genome of Papilio glaucus and its phylogenetic implications. Meta Gene. 5:68–83.
- Shen J, Cong Q, Grishin NV. 2016. The complete mitogenome of Achalarus lyciades (Lepidoptera: Hesperiidae). Mitochondrial DNA B Resour. 1:581–583.
- Stallings DB, Turner JR, Stallings VN. 1961. A new subspecies of Agathymus mariae from Mexico (Megathymidae). J Lep Soc. 15:19–22.
- Zhang J, Cong Q, Fan XL, Wang R, Wang M, Grishin NV. 2017b. Mitogenomes of Giant-Skipper Butterflies reveal an ancient split between deep and shallow root feeders. F1000Res. 6:222.
- Zhang J, Cong Q, Shen J, Fan XL, Wang M, Grishin NV. 2017a. The complete mitogenome of Euschemon rafflesia (Lepidoptera: Hesperiidae). Mitochondrial DNA B Resour. 2:136–138.
- Zhang J, Cong Q, Shen J, Wang R, Grishin NV. 2017c. The complete mitochondrial genome of a skipper Burara striata (Lepidoptera: Hesperiidae). Mitochondrial DNA B Resour. 2:145–147.
