Abstract
Complete mitogenome sequence for eastern Asian population of Cheilonereis cyclurus (Polychaeta: Nereididae) was determined for the first time. The length of circular genome of C. cyclurus is 14,917 bp including 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and a non-coding region of 383 bp. The gene order of C. cyclurus is identical to that of the following four nereidid species: Hediste diadroma, Namalycastis abiuma, Paraleonnates uschakovi, Tylorrhynchus heterochaetus. The phylogenetic position of C. cyclurus compared to 16 selected polychaetes was conducted and present species is closely related to the clade containing Perinereis nuntia, P. aibuhitensis, and Platynereis dumerilii with high bootstrap value.
The genus Cheilonereis Benham (Citation1916) belongs to family Nereididae Blainville (Citation1818) and has two nominal species: C. cyclurus (Harrington Citation1897) and C. peristomialis Benham (Citation1916) (Bakken and Wilson Citation2005; Read and Fauchald Citation2017). They are commensal with hermit crabs. Of these, C. cyclurus is widely distributed in the east and west coasts of the north Pacific (Harrington Citation1897; Berkeley and Berkeley Citation1948; Imajima Citation1972; Paik Citation1977). This species is easily distinguishable among nereidids with unique characteristic of ventral peristomial flap at the oral ring. However, recent phylogenetic study using morphological characteristics revealed an ambiguous relationship with Perinereis species (Bakken and Wilson Citation2005). Also, the comparison of partial mitochondrial COI sequences revealed significant difference between east and west north Pacific populations (; personal data of the author). Although, the comparison of complete mitogenome sequence may provide more valuable evolutionary information for phylogenetic studies (Chen et al. Citation2016), only seven nereidids are available (Boore Citation2001; Won et al. Citation2013; Kim et al. Citation2014; Chen et al. Citation2016; Kim et al. Citation2016; Lin et al. Citation2016; Park et al. Citation2016). The purpose of this study was to determine the complete mitogenome sequence for eastern Asian population of C. cyclurus.
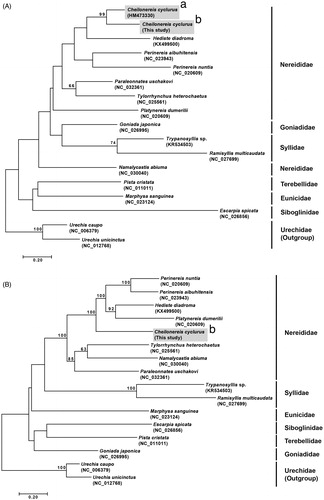
Figure 1. Maximum-likelihood (ML) trees constructed using MEGA 7.0 software. Bootstrap replicates were performed 1000 times. Bootstrap values above 60% were indicated on the cladogram. (A) ML tree based on 16 mitogenome sequences including Cheilonereis cyclurus (this study). (B) ML tree based on 17 partial mitochondrial cytochrome c oxidase subunit I (COI) including C. cyclurus from the Northeast and Northwest Pacific. (a) Specimen from Bamfield, British Columbia, Canada (Carr et al. Citation2011); (b) specimen from Goseong-gun, Gangwon-do, Korea.
The specimen was collected in the subtidal zone (about 200 m depth) of the East Sea near Goseong-gun, Gangwon-do, Korea with local fishermen’s gill net. A live specimen was used to extract pure mitochondrial genomic DNA. Total mt DNA was extracted using the DNesy Blood & Tissue Kit (Qiagen) according to the manufacturer’s protocol. mt DNA was amplified using REPLI-g mitochondrial DNA kit (Qiagen) to increase concentration for NGS analysis. A voucher specimen was housed at the National Institute of Biological Resources (NIBRIV0000787920).
Total length of the C. cyclurus complete mitogenome was 14,917 bp (GenBank accession no. MF538532), and encoded 37 genes (13 protein-coding genes, two rRNA genes, and 22 tRNA genes) and a non-coding region of 383 bp. According to Park et al. (Citation2016), nereidids have two types of gene order groups depending on the patterns of following regions: tRNA-Tyr, ATP8, tRNA-Met, and tRNA-Asp (Group 1) and tRNA-Met, tRNA-Asp, ATP8, and tRNA-Tyr (Group 2). Gene order of C. cyclurus is identical to Group 1. The initiation codons of genes include ATT (nad1) and ATG (cox1–3, nad6, cytb, nad5, nad4L, nad2–4, atp6, atp8). Stop codons include TAA (cox2, cytb, nad2–4, nad4L, atp6) except some genes (cox1, cox3, nad1, nad5, nad6, atp8) terminated with T.
To examine the phylogenetic position of C. cyclurus, phylogenetic analyses were conducted using MEGA7 (Kumar et al. Citation2016) and reconstructed the tree in maximum-likelihood method by concatenated 13 protein coding genes from 16 selected polychaetes. As a result, C. cyclurus was grouped into family Nereididae and closely related to the clade containing Perinereis nuntia, P. aibuhitensis, and Platynereis dumerilii with high bootstrap value ().
The new mitogenome sequence determined from this study will be useful for further taxonomic and phylogenetic examination not only for eastern Asian population of C. cyclurus but also other nereidid species.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and the writing of this study.
Additional information
Funding
References
- Bakken T, Wilson RS. 2005. Phylogeny of nereidids (Polychaeta, Nereididae) with paragnaths. Zool Scripta. 34:507–547.
- Benham WB. 1916. Report on the Polychaeta obtained by the F.I.S. ''Endeavour'' on the coasts of New South Wales, Victoria, Tasmania and South Australia. Fisheries: biological results of the fishing experiments carried on by the F.I.S. ''Endeavour'', 1909–14. H.C. Dannevig; Sydney, 4. p. 125–162, plates XLVI–XLVIII.
- Berkeley E, Berkeley C. 1948. Canadian Pacific Fauna. 9 Annelida. 9b (1). Polychaeta Errantia. Toronto: Fisheries Research Board of Canada; p. 1–100.
- Blainville H. 1818. Mémoire sur la classe des Sétipodes, partie des Vers à sang rouge de M. Cuvier, et des Annélides de M. de Lamarck. Bull Des Sci Par La Société Philomatique De Paris. 1818:78–85.
- Boore JL. 2001. Complete mitochondrial genome sequence of the polychaete annelid Platynereis dumerilii. Mol Biol Evol. 18:1413–1416.
- Carr CM, Hardy SM, Brown TM, Macdonald TA, Hebert PDN. 2011. A tri-oceanic perspective: DNA barcoding reveals geographic structure and cryptic diversity in Canadian polychaetes. PLoS One. 6:e22232.
- Chen X, Li M, Liu H, Li B, Guo L, Meng Z, Lin H. 2016. Mitochondrial genome of the polychaete Tylorrhynchus heterochaetus (Phyllodocida, Nereididae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:3372–3373.
- Harrington NR. 1897. On nereids commensal with hermit crabs. Trans N Y Acad Sci. 16:214–222. plates XVI–XVIII.
- Imajima M. 1972. Review of the annelid worms of the family Nereidae of Japan, with descriptions of five new species or subspecies. Bull Natl Sci Mus Tokyo. 15:37–153.
- Kim H, Jung G, Lee YC, Pae SJ, Kim CG, Lee YH. 2014. The complete mitochondrial genome of the marine polychaete: Perinereis aibuhitensis (Phyllodocida, Nereididae). Mitochondrial DNA. 26:869–870.
- Kim H, Kim HJ, Lee YH. 2016. The complete mitochondrial genome of the marine polychaete: Hediste diadroma (Phyllodocida, Nereididae). Mitochondrial DNA B Resour. 1:822–823.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lin GM, Shen KN, Hsiao CD. 2016. Next generation sequencing yields the complete mitogenome of nereid worm, Namalycastis abiuma (Annelida: Nereididae). Mitochondrial DNA B Resour. 1:103–104.
- Paik E. 1977. Studies of polychaetous annelid worms of the family Nereidae in Korea. Res Bul Hyosung Women Coll. 19:131–227.
- Park T, Lee SH, Kim W. 2016. Complete mitochondrial genome sequence of the giant mud worm Paraleonnates uschakovi Khlebovich & Wu, 1962 (Polychaeta: Nereididae). Mitochondrial DNA B Resour. 1:640–642.
- Read G, Fauchald K, editor. 2017. World Polychaeta database. [accessed 2017-08-01]. http://www.marinespecies.org/polychaeta.
- Won EJ, Rhee JS, Shin KH, Lee JS. 2013. Complete mitochondrial genome of the marine polychaete, Perinereis nuntia (Polychaeta, Nereididae). Mitochondrial DNA. 24:342–343.
