Abstract
The caddisfly Anabolia bimaculata, is a rapidly-maturing caddisfly found in temporary ponds across western North America. Whole genome Illumina sequencing facilitated the assembly of a complete circular mitochondrial genome of 15,048 bp from A. bimaculata consisting of 78.2% AT nucleotides, 22 tRNAs, 13 protein-coding genes, two rRNAs and a control region arranged in the canonical and ancestral gene order found in insects. This gene order is consistent with the gene order found in most other caddisfly species. Maximum likelihood phylogenetic reconstruction places A. bimaculata within a monophyletic Order Trichoptera and sister to the primitive lepidopteran Micropterix calthella (Micropterigidae).
Aquatic caddisfly larvae (Insect Order Trichoptera) are excellent habitat quality bioindicators (Wissinger et al. Citation2003), but can be challenging to identify, so molecular tools for taxonomic identification are valuable (Ruiter et al. Citation2013). Here we report the first complete mitogenome for a New World caddisfly.
On 17–18 July 2015, a USDA blacklight trap (Winter Citation2000) was deployed to collect night-flying insects at the Living Prairie Museum (GPS 49.889607 N, −97.270487 W), 12.9 ha of relict prairie habitat in Winnipeg, Manitoba, Canada (Living Prairie Mitogenomics Consortium Citation2017). One adult Anabolia bimaculata (Limnephilidae), a rapidly-maturing caddisfly found in temporary ponds across western North America (Berté and Pritchard Citation1986), was trapped (specimen number 2015.07.17.018). The specimen was pinned and deposited in the Wallis Roughley Museum of Entomology, University of Manitoba (voucher JBWM0362999).
DNA was prepared (McCullagh and Marcus Citation2015) and sequenced by Illumina MiSeq (San Diego, CA) (Peters and Marcus Citation2017). The complete mitogenome for A. bimaculata (Genbank MF680449) was assembled by Geneious 10.1.2 from 8,292,766 paired 75 bp reads (total 0.63 Gb) using a Eubasilissa regina (Trichoptera: Phryganeidae) reference mitogenome (NC_023374.1) (Wang et al. Citation2014). Annotation was performed with reference to E. regina and Hydropsyche pellucidula (Trichoptera: Hydropsychidae, KT876876.1) mitogenomes (Linard et al. Citation2017). The complete A. bimaculata nuclear rRNA repeat (Genbank MF680448) was also assembled using a Stenopsyche marmorata reference sequence (Trichoptera: Stenopsychidae, LC094265.1) and annotated with respect to S. marmorata and Attacus ricini (Lepidoptera: Saturniidae, AF463459) repeats (Wang et al. Citation2003).
The circular 15,048 bp mitogenome assembly of A. bimaculata was made from 40,856 paired reads with nucleotide composition of 39.1% A, 14.1% C, 8.5% G, and 38.1% T. Anabolia bimaculata maintains complete synteny with nearly all other sequenced trichopteran mitogenomes and shows the canonical insect mitochondrial gene order. It lacks the rearrangements previously reported in the H. pellucidula mitogenome (Linard et al. Citation2017). Anabolia bimaculata COX1 features an aberrant start codon (CGA) and three mitochondrial protein-coding genes (ATP8, NAD1 NAD4) have single-nucleotide (T) stop codons that are completed by the post-transcriptional addition of 3′ A residues. Anabolia bimaculata tRNAs have standard cloverleaf secondary structures except for trnS (AGN) which has the dihydrouridine arm replaced by a loop as determined by analysis in Mfold (Zuker Citation2003). The rRNAs (789 bp 12S and 1354 bp 16s) are composed of 84.7% AT while the putative control region (276 bp) is 82.8% AT.
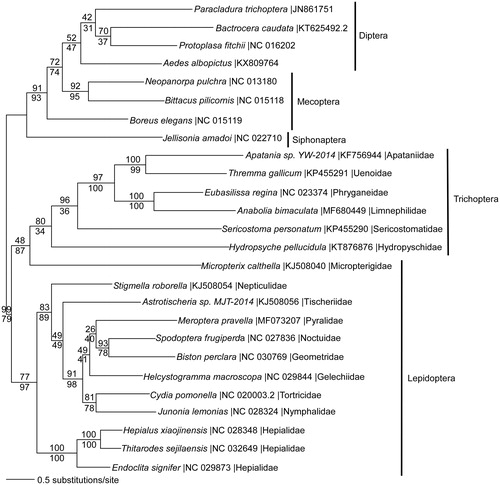
We reconstructed a phylogeny using 13 protein coding genes from mitogenomes of A. bimaculata, five other trichopteran species, and representatives from related holometabolous insect orders. DNA sequences from each gene were aligned in CLUSTAL Omega (Sievers et al. Citation2011), concatenated, and analysed by maximum likelihood (ML) and parsimony in PAUP* 4.0b8/4.0d78 (Swofford Citation2002) (). ML phylogenetic analysis places A. bimaculata within a monophyletic Trichoptera, sister to the primitive lepidopteran Micropterix calthella (Micropterigidae). The Trichoptera + Micropterix clade is sister to the remaining Lepidoptera. This unconventional placement may be due to missing data in Micropterix. The arrangement of taxa within the Trichoptera is consistent with prior published reports (Linard et al. Citation2017).
Figure 1. Maximum likelihood phylogeny (GTR + I + G model, I = 0.2790, G = 0.4760, likelihood score 153494.68624) of Anabolia bimaculata and other Trichoptera species with representatives from the sister-order Lepidoptera (moths and butterflies), and from Diptera (flies), Mecoptera (scorpionfiles), and Siphonaptera (fleas) based on 1 million random addition heuristic search replicates (with tree bisection and reconnection) of mitochondrial protein coding genes. One million maximum parsimony heuristic search replicates produced a single tree (37,008 steps) with nearly identical tree topology except that Micropterix is the sister taxon to Hydropsyche, rather than to the entire trichopteran clade. Numbers above each node are maximum likelihood bootstrap values and numbers below each node are maximum parsimony bootstrap values (each from 1 million random fast addition search replicates).
Acknowledgements
We thank Sarah Semmler and Kyle Lucyk for permitting and encouraging our work at the Living Prairie Museum. We thank Melissa Peters for help with fieldwork, Melanie Lalonde for assistance in the lab, and Aleksandar Ilik and Debbie Tsuyuki (Children’s Hospital Research Institute of Manitoba Next Generation Sequencing Platform) for assistance with library preparation and sequencing.
Disclosure statement
The authors report no conflicts of interest, and are solely responsible for this paper.
Additional information
Funding
References
- Berté SB, Pritchard G. 1986. The life histories of Limnephilus externus Hagen Anabolia bimaculata (Walker), and Nemotaulius hostilis (Hagen) (Trichoptera, Limnephilidae) in a pond in southern Alberta, Canada. Can J Zool. 64:2348–2356.
- Linard B, Arribas P, Andujar C, Crampton-Platt A, Vogler AP. 2017. The mitogenome of Hydropsyche pellucidula (Hydropsychidae): first gene arrangement in the insect order Trichoptera. Mitochondrial DNA A. 28:71–72.
- Living Prairie Mitogenomics Consortium. 2017. The complete mitochondrial genome of the lesser aspen webworm moth Meroptera pravella (Insecta: Lepidoptera: Pyralidae). Mitochondrial DNA B Resour. 2:344–346.
- McCullagh BS, Marcus JM. 2015. The complete mitochondrional genome of Lemon Pansy, Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae). J Asia-Pacific Ent. 18:749–755.
- Peters MJ, Marcus JM. 2017. Taxonomy as a hypothesis: testing the status of the Bermuda buckeye butterfly Junonia coenia bergi (Lepidoptera: Nymphalidae). Syst Ent. 42:288–300.
- Ruiter DE, Boyle EE, Zhou X. 2013. DNA barcoding facilitates associations and diagnoses for Trichoptera larvae of the Churchill (Manitoba, Canada) area. BMC Ecol. 13:5.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.
- Swofford DL. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Massachusetts, USA: Sinauer Associates, Sunderland.
- Wang SQ, Zhao MJ, Li TP. 2003. Complete sequence of the 10.3 kb silkworm Attacus ricini rDNA repeat, determination of the transcriptional initiation site and functional analysis of the intergenic spacer. DNA Seq. 14:95–101.
- Wang Y, Liu X, Yang D. 2014. The first mitochondrial genome for caddisfly (Insecta: Trichoptera) with phylogenetic implications. Int J Biol Sci. 10:53–63.
- Winter WD. 2000. Basic techniques for observing and studying moths and butterflies. Vol. 5. Los Angeles (CA): The Lepidopterists' Society.
- Wissinger SA, Brown WS, Jannot JE. 2003. Caddisfly life histories along permanence gradients in high-altitude wetlands in Colorado (USA). Freshwater Biol. 48:255–270.
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415.
