Abstract
The complete mitochondrial genome of Phlebopus portentosus was determined using Illumina sequencing. The circular genome is 42,963 bp in length with GC content of 21.37%. It contains 14 putative protein-coding genes, the ribosomal RNA subunits and 23 tRNAs, all located on the same strand. The evolutionary relationships between P. portentosus and other 23 representative basidiomycete species were revealed based on sequences at the 14 concatenated mitochondrial protein-coding genes.
Phlebopus portentosus is an artificially cultivatable boletus mushroom and the mitochondrion is an important essential organelle in mushrooms (Burger et al. Citation2003; Ji et al. Citation2011). Elucidating the structure and function of P. portentosus mitogenome is important for understanding its genetics, diversity and evolution. The genome sequence of P. portentosus has been reported (Cao et al. Citation2015). However, little is known about its mitogenome. Here, we report the mitogenome of P. portentosus and investigate the phylogenetic relationships of P. portentosus and other related species based on mitochondrial protein-coding genes.
The mitogenome sequence of P. portentosus was obtained from strain PP33 from the Institute of Tropical Crops of Yunnan Province (collected from Dongfeng Farmland which is 34 km south to Jinghong City, Xishuangbanna Autonomous Prefecture, Yunnan Province, China. N 21°42’ E 100°45. And the single spore isolate of P. portentosus was deposited as living cultures in China General Microbiological Culture Collection Center under the accession number: CGMCC 6240). The published mitogenome of Ganoderma lucidumd (GenBank ID: KP410262) was used as a reference. The mitogenome sequence of P. portentosus was identified by BLAST as scaffold54 (GenBank ID: KN880405) in the whole genome assembly and annotated using MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) and GLIMMER (https://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi). tRNAs were annotated using tRNAscan-SE (Schattner et al. Citation2005). All ORFs were searched and identified by ORFFinder.
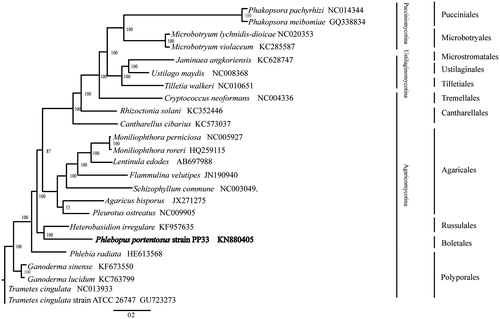
The mitogenome of strain PP33 is a typical circular DNA molecule of 42,963 bp in length with a GC content of 21.37%. Genome annotation identified ORFs that encode 14 conserved proteins, 23 tRNAs, 2 homing endonucleases, the small ribosomal RNAs (rns) and the large ribosomal RNA (rnl). The 14 conserved proteins include 7 subunits of NAD dehydrogenase (nad1-6 and nad4L genes), 3 cytochrome oxidases (cox1-3), apocyto chrome b (cob) and 3 subunits of ATP synthase (atp6, apt 8 and apt 9). The 23 tRNA genes covered all 20 standard amino acids. There were 5 introns distributed in this mitogenome, 4 of which were found in the cox1 gene: two were group I and the other two were group II. Phylogenetic analysis based on concatenated sequences of 14 conserved mitochondrial protein-coding genes of 23 Basidiomycetes was performed by Bayesian inference (BI). As shown in . P. portentosus was a member of Agaricomycotina and closely related to Russulales and Polyporales. The inferred relationships using mitochondrial gene sequences were the same as those constructed based on sequences of nuclear genes (Garcia-Sandoval et al. Citation2011; Wu et al. Citation2014). The Genbank Accession number for Phlebopus Portentosus is MK571437.
Figure 1. Phylogenetic relationships among 23 basidiomycete fungi inferred based on the concatenated amino acid sequences of 14 mitochondrial protein-coding genes. The 14 mitochondrial protein-coding genes were: nad1, nad2, nad3, nad4, nad4L, nad5, nad6, cox1, cox2, cox3, cob, atp6, atp8, atp9. The tree was generated using Bayesian inference (BI). Numerical values along branches represent statistical support based on 1000 randomizations.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the manuscript.
Additional information
Funding
References
- Burger G, Gray MW, Lang BF. 2003. Mitochondrial genomes: anything goes. Trends Genet. 19:709.
- Cao Y, Zhang Y, Yu Z, Wang P, Tang X, He X, Mi F, Liu C, Yang D, Xu J. 2015. Genome sequence of Phlebopus portentosus strain PP33, a cultivated bolete. Genome Announc. 3:e00326-15.
- Garcia-Sandoval R, Wang Z, Binder M, Hibbett DS. 2011. Molecular phylogenetics of the Gloeophyllales and relative ages of clades of Agaricomycotina producing a brown rot. Mycologia. 103:510–524.
- Ji KP, Cao Y, Zhang CX, He MX, Liu J, Wang WB, Wang Y. 2011. Cultivation of Phlebopus portentosus, in southern China. Mycological Progress. 10:293–300.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:(Web Server issue) W686.
- Wu G, Feng B, Xu J, Zhu XT, Li YC, Zeng NK, Hosen MI, Yang ZL. 2014. Molecular phylogenetic analyses redefine seven major clades and reveal 22 new generic clades in the fungal family Boletaceae. Fungal Diversity. 69: 93–115.
