Abstract
In the present study, the complete mitogenome sequence of Culter oxycephaloides was determined using PCR amplification and DNA sequencing, which contains 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a non-coding control region with the total length of 16,619 bp. Except for eight tRNA and ND6 genes, all other mitochondrial genes are encoded on the heavy strand. The codon usage followed the typical vertebrate mitochondrial pattern (ATG or GTG for start codon and TAA or TAG for stop codon). There are 12 regions of gene share totalling 55 bp and 11 intergenic spacer regions totalling 84 bp. The complete mitochondrial genome sequence is useful for phylogenetic analysis and studies of population genetics of C. oxycephaloides.
Here, one Culter oxycephaloides was caught in Shashi section of middle reach in Yangtze River, Hubei province in China. Total genomic DNA from fin tissue was extracted using a modified ammonium acetate precipitation protocol (Nicholls et al. Citation2000) and stored at −20 °C. The rest of the specimens were kept in a laboratory of Yangtze River Fisheries Research Institute, Chinese Academy of Fishery Sciences. We amplified and sequenced the whole mitochondrial genome sequence of C. oxycephaloides with 16 pairs of primers designed in accordance with equivalent mitochondrial genome sequences of 30 species from Cypriniformes (Wang et al. Citation2011). The DNA sequence with a Web-based tool DOGMA (Wyman et al. Citation2004) was annotated, which has ever been used in many previous studies. Here we presented the complete mitochondrial genome of C. oxycephaloides that was deposited in GenBank with the accession number KY404014.
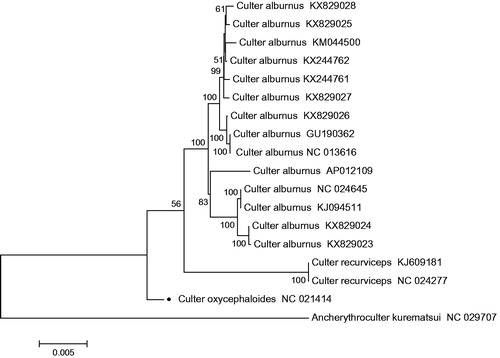
The complete mitochondrial genome of C. oxycephaloides was found to be similar to that of most other vertebrates (Huang et al. Citation2012). It was sequenced and determined to be 16,619 bp in length, including 13 typical vertebrate protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a control region. The majority of the mitochondrial genes of C. oxycephaloides are encoded on heavy strand (H-strand) except for ND6 and eight tRNA genes(tRNA-Gln, Ala, Asn, Cys, Tyr, Ser (UCN), Glu, and Pro), which are encoded on light strand (L-strand). The base composition of H-strand was as follows 31.3% A, 27.88% C, 24.75% T, 16.07% G and AT content was 56.06%. The relative order of nucleotide composition matches with the nucleotide pattern of other cyprinids in the order: A > C > T > G (Wang Citation2008). The GC content of cyprinid mitochondrial genomes is always lower than AT content and this is also evident in other teleost species (Yue et al. Citation2006). Of the 13 protein-coding genes, gene overlaps can be observed between four pairs of the contiguous genes, ATP8–ATP6, ATP6–COIII, ND4L–ND4, and ND5–ND6, and they overlap by seven, one, seven, and four nucleotides, respectively and 84 bp intergenic nucleotides in the whole genome, ranging from 1 to 31bp. All 13 protein-coding genes of C. oxycephaloides share the start codon ATG, except for COI, which begins with GTG. Open reading-frames of five protein-coding genes end with TAA and one protein-coding genes end with TAG, while, the others with incomplete stop codons, TA- (COIII, ND4) or T- (ND2, COII, ND3, ND6 and Cyt b). To further validate the new sequences, MEGA 5 (Tamura et al. Citation2011) was used to produce the phylogenetic tree of C. oxycephaloides and all other Culter sequences in NCBI based on the Neighbour-Joining method (), with Ancherythroculter kurematsui as an outgroup. As expceted, C. oxycephaloides clustered with the other two Culter species, C. alburnus and C. recurviceps, and with Ancherythroculter kurematsui on different branches.
Figure 1. N-J phylogenetic tree of C. oxycephaloides and all other Culter sequences in NCBI, with Ancherythroculter kurematsui as an outgroup. The round dot indicate the individual sampled in this study.
In this study, we reported the complete mitochondrial genome (mitogenome) of C. oxycephaloides and hope it may play important roles in the studies of population genetics and phylogenetic analysis of this species.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Huang Y, Zhao G, Peng Z. 2012. Mitochondrial genome of Onychostoma lini (Teleostei, Cypriniformes). Mitochondrial DNA. 23:173–175.
- Nicholls JA, Double MC, Rowell DM, Magrath D. 2000. The evolution of cooperative and pair breeding in thornbills Acanthiza (Pardalotidae). J Avian Biol. 31:165–176.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. Mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Wang C, Chen Q, Lu G, Xu J, Yang Q, Li S. 2008. Complete mitochondrial genome of the grass carp (Ctenopharyngodon idella, Teleostei): insight into its phylogenic position within Cyprinidae. Gene. 424:96–101.
- Wang J, Li P, Zhang Y, Peng Z. 2011. The complete mitochondrial genome of Chinese rare minnow, Gobiocypris rarus (Teleostei: Cypriniformes). Mitochondrial DNA. 22:178–180.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yue GH, Liew WC, Orban L. 2006. The complete mitochondrial genome of a basal teleost, the Asian arowana (Scleropages formosus, Osteoglossidae). BMC Genomics. 7:1–13.
