Abstract
The complete plastome sequence of Garcinia mangostana L. (Clusiaceae) is completed in this study (NCBI acc. no. KX822787). This is a first complete plastome sequence from the Clusiaceae. The complete plastome size is 158,179 bp in length and consists of a large single copy of 86,458 bp and a small single copy of 17,703 bp, separated by two inverted repeats of 27,009 bp. The G. mangostana plastome shows four minor structural modifications including infA gene loss, rpl32 gene loss, ycf3 gene intron loss and a 363 bp inversion between trnV-UAC and atpE gene. The plastome contains 111 genes, of which 77 are protein-coding genes, 30 are tRNA genes and four are rRNA genes. The average A-T content of the plastome is 63.9%. A total of 110 simple sequence loci are identified from the genome. Phylogenetic analysis reveals that G. mangostana is a sister group of Erythroxylum novogranatense (Erythroxylaceae) with 78% bootstrap support.
Garcinia mangostana L. is commonly known as ‘the queen of fruits’ or mangosteen. It is one of the most delicious tropical fruits originated in Malaysia and Indonesia (Kim Citation2011). It belongs to the family Clusiaceae of Malpighiales (APG IV Citation2016). The family Clusiaceae consists of 13 genera and approximately 750 species including many fruit crops (Christenhusz and Byng Citation2016). But, the complete plastome sequence is not known from the family. Therefore, we first report the complete plastome of G. mangostana (Clusiaceae) in this study. It will be a useful reference for the phylogenetic and evolutionary studies of Malphigiales.
The leaves of G. mangostana used in this study were collected from the Korea University greenhouse, where we grew the plants from seeds originally collected from Thailand. A voucher specimen was deposited in the Korea University Herbarium (KUS acc. no. 2014-0243). Fresh leaves were ground into powder in liquid nitrogen and total DNAs were extracted using the cetyl trimethyl ammonium bromide (CTAB) method (Doyle and Doyle Citation1987). The DNAs were further purified by the ultracentrifugation and dialysis (Palmer Citation1986). The genomic DNAs are deposited in the Plant DNA Bank in Korea (PDBK acc. no. 2014-0243). The complete plastome sequence was generated using an Illumina HiSeq 2000 system (Illumina Inc., San Diego, CA). An average coverage of sequences was 346 times of its annotated plastome size. Annotations were performed using the National Center for Biotechnology Information (NCBI) BLAST and tRNAscan-SE programmes (Lowe and Eddy Citation1997).
The gene order and structure of the G. mangostana plastome are similar to those of a typical angiosperm (Shinozaki et al. Citation1986; Kim and Lee Citation2004; Yi and Kim Citation2012) except four minor modifications. The infA and rpl32 genes were lost in the G. mangostana plastome. The two gene losses were also reported from other Malpighialian families (Tangphatsornruang et al. Citation2011; Huang et al. Citation2014; Bardon et al. Citation2016; Cheon et al. Citation2017). The ycf3 gene usually consists of three exons and two introns in angiosperm. But, the second intron was lost in G. mangostana plastome. The G. mangostana plastome had a small 363 bp inversion between trnV-UAC and atpE region. The palindromic repeats of 15 bp (ACATCCTATTTCTTT/AAAGAAATAGGATGT) were located on the two break points of inversion. This is a new inversion report, but there are several reports of other small inversions due to palindromic repeats (Kim and Lee Citation2004; Catalano et al. Citation2009).
The complete plastome is 158,179 bp in length and consists of a large single copy (LSC) region of 86,458 bp and a small single copy (SSC) region of 17,703 bp separated by two inverted repeats (IR) of 27,009 bp. The plastome comprises 111 unique genes (77 protein-coding genes, 30 tRNA genes and four rRNA genes). Seven protein-coding, seven tRNA and four rRNA genes are duplicated in the IR regions. The average A-T content of the plastome is 63.9%, whereas that in the LSC, SSC and IR regions is 66.5%, 69.8% and 57.8%, respectively. Seventeen genes have one intron and one gene (clpP) has two introns. A total of 110 simple sequence repeat (SSR) loci are scattered among the plastome. Among these, 88, 15 and 7 are mono-SSR, di-SSR and tri-SSR loci, respectively.
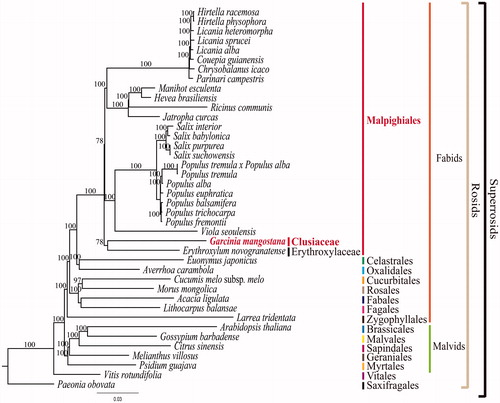
To validate the phylogenetic relationships of G. mangostana in rosids, we constructed a maximum likelihood (ML) tree by using 40 super-rosids taxa. Phylogenetic analysis was performed on a data set that included 78 protein-coding genes and four rRNA genes using RAxML version 7.7.1 (Stamatakis et al. Citation2008). The 82 gene sequences (77,445 bp in length) were aligned with the MUSCLE programme using Geneious version 6.1.8 (Biomatters Ltd.; Kearse et al. Citation2012). As a result, G. mangostana forms a clade with Erythroxylum novogranatense (Erythroxylaceae) with a 78% bootstrap value (). In previous phylogenetic studies, Clusiaceae belongs to the clusioids group and has been known to be the closest relationship to Bonnetiaceae (Wurdack and Davis Citation2009; Ruhfel et al. Citation2011; Xi et al. Citation2012). There are 36 families in Malphigiales. But, the complete plastomes were reported from only six families so far. In order to solidify the interfamilial relationships in Malphigiales, we need a complete plastome sequences from other families.
Figure 1. Maximum Likelihood (ML) tree based on 78 protein-coding and four rRNA genes from 40 plastomes as determined by RAxML(−ln L = −466117.224833). The numbers at each node indicate the ML bootstrap values. GenBank accession numbers of taxa are shown below, Acacia ligulata (NC_026134), Arabidopsis thaliana (NC_000932), Averrhoa carambola (NC_033350), Chrysobalanus icaco (NC_024061), Citrus sinensis (NC_008334), Couepia guianensis (NC_024063), Cucumis melo subsp. melo (NC_015983), E. novogranatense (NC_030601), Euonymus japonicus (NC_028067), G. mangostana (KX822787), Gossypium barbadense (NC_008641), Hevea brasiliensis (NC_015308), Hirtella physophora (NC_024066), H. racemose (NC_024060), Jatropha curcas (NC_012224), Larrea tridentata (NC_028023), Licania alba (NC_024064), L. heteromorpha (NC_024062), L. sprucei (NC_024065), Lithocarpus balansae (NC_026577), Manihot esculenta (NC_010433), Morus mongolica (NC_025772), Melianthus villosus (NC_023256), Paeonia obovata (NC_026076), Parinari compestris (NC_024067), Populus alba (NC_008235), P. balsmifera (NC_024735), P. euphratica (NC_024747), P. fremontii (NC_024734), P. tremula (NC_024725), P. tremula × P. alba (NC_028504), P. trichocarpa (NC_009143), Psidium guajava (NC_033355), Ricinus communis (NC_016736), Salix babylonica (NC_028350), S. interior (NC_024681), S. purpurea (NC_026722), S. suchowensis (NC_026462) Viola seoulensis (NC_026986) and Vitis rotundifolia (NC_023790).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- APG IV. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Bardon L, Sothers C, Prance GT, Malé PJG, Xi Z, Davis CC, Murienne J, García-Villacorta R, Coissac E, Lavergne S. 2016. Unraveling the biogeographical history of Chrysobalanaceae from plastid genomes. Am J Bot. 103:1089–1102.
- Catalano SA, Saidman BO, Vilardi JC. 2009. Evolution of small inversions in chloroplast genome: a case study from a recurrent inversion in angiosperms. Cladistics. 25:93–104.
- Cheon KS, Yang JC, Kim KA, Jang SK, Yoo KO. 2017. The first complete chloroplast genome sequence from Violaceae (Viola seoulensis). Mitochondrial DNA A DNA Mapp Seq Anal. 28:67–68.
- Christenhusz MJ, Byng JW. 2016. The number of known plants species in the world and its annual increase. Phytotaxa. 261:201–217.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Hefer CA, Kolosova N, Douglas CJ, Cronk QC. 2014. Whole plastome sequencing reveals deep plastid divergence and cytonuclear discordance between closely related balsam poplars, Populus balsamifera and P. trichocarpa (Salicaceae). New Phytol. 204:693–703.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kim K-J. 2011. Tropical fruit resources. Seoul: Geobook.
- Kim K-J, Lee HL. 2004. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 11:247–261.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Palmer JD. 1986. Isolation and structural analysis of chloroplast DNA. Methods Enzymol. 118:167–186.
- Ruhfel BR, Bittrich V, Bove CP, Gustafsson MH, Philbrick CT, Rutishauser R, Xi Z, Davis CC. 2011. Phylogeny of the clusioid clade (Malpighiales): evidence from the plastid and mitochondrial genomes. Am J Bot. 98:306–325.
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 5:2043–2049.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771.
- Tangphatsornruang S, Uthaipaisanwong P, Sangsrakru D, Chanprasert J, Yoocha T, Jomchai N, Tragoonrung S. 2011. Characterization of the complete chloroplast genome of Hevea brasiliensis reveals genome rearrangement, RNA editing sites and phylogenetic relationships. Gene. 475:104–112.
- Wurdack KJ, Davis CC. 2009. Malpighiales phylogenetics: gaining ground on one of the most recalcitrant clades in the angiosperm tree of life. Am J Bot. 96:1551–1570.
- Xi Z, Ruhfel BR, Schaefer H, Amorim AM, Sugumaran M, Wurdack KJ, Endress PK, Mattews ML, Stevens PF, Davis CC. 2012. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc Natl Acad Sci USA. 109:17519–17524.
- Yi D-K, Kim K-J. 2012. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS One. 7:e35872.
