Abstract
Agrocybe aegerita is a medicinally and nutritionally important edible basidiomycete. Despite previous phylogenetic studies, the taxonomy of A. aegerita complex remains unclear due to lacking of resolutive data. Herein, the complete mitochondrial genome of A. aegerita is reported and analyzed. The mitogenome length was 116,329 bp, with a GC content of 27.6%, include 17 typical protein-coding genes, two ribosomal protein genes (rps3), two ribosomal RNA genes and a set of 32 transfer RNA genes. A phylogenetic analyses using complete mitogenome in Agaricales showed that A. aegerita is closely related to the genus Pleurotus and represents a clade clearly independent from other Agaricales species.
Agrocybe aegerita (Basidiomycota, Agaricomycetes, Agaricales, Strophariaceae) (Matheny et al. Citation2006; Uhart et al. Citation2007) is used as a nutritious food and herbal medicinal around the world (Wasser and Weis Citation1999; Zhao et al. Citation2003; Tsai et al. Citation2006; Thushara et al. Citation2008). Agrocybe aegerita is cultivated and is a biological model for the developmental genetic engineering (Barroso et al. Citation1995; Noël and Labarère Citation1994). Agrocybe aegerita has been characterized as a multispecies complex due to a wide range of variation of morphological and physiological characters, the disordered designation has hindered the development of breeding programs (Singer Citation1986; Uhart et al. Citation2007; Chen et al. Citation2012). Using rDNA and ITS sequence in A. aegerita complex showed a high diversity in the species/varieties collected from different place (Uhart et al. Citation2007; Chen et al. Citation2012). However, the taxonomical status of this species remains unclear, and no complete mitogenome is available to date. Here, we report the complete mitochondrial genome of A. aegerita to provide new genetic resources and to uncover the taxonomical status of this species.
The strain SWS_17 of A. aegerita is conserved in the Biology Institute, Guangxi Academy of Sciences (Nanning, Guangxi, PR China). Total genomic DNA was extracted from an individual previously grown on Potato Dextrose Agar (PDA) medium at 25 °C for 2 weeks, using the Promega Genomic DNA Purification System (Promega, Madison, WI). Library construction and sequencing were processed by Novogene (Beijing, China), according to the Illumina HiSeqX-ten system manufacturer (Illumina Inc., San Diego, CA) instructions. The complete mitogenome was de novo assembled using org.asm v0.2.05 (available at http://pythonhosted.org/ORG.asm/) followed by manual checking in Geneious R9 v9.1.6 (Biomatters Ltd, Auckland, New Zealand) as described previously (Hinsinger and Strijk Citation2016; Jiang et al. Citation2016). Genome annotation was performed with the on line program DOGMA (Wyman et al. Citation2004) and manually checked in Geneious. The complete mitogenome sequence of A. aegerita was submitted to GenBank under accession no. MF979820. Phylogenetic analysis was performed with Phyml 3.1 to construct a maximum-likelihood (ML) tree including 10 mitochondrions available of Agaricales in GenBank (see ). Nucleotide substitution models were tested by jmodeltest (Posada Citation2009).
Figure 1. ML phylogenetic tree of the 10 available mitochondrial sequences of Agaricales in GenBank, plus the mitochondrial sequence of Agrocybe aegerita. The tree is rooted with Heterobasidion irregulare. Bootstraps values (1000 replicates) are shown at the nodes. Scale in substitution per site.
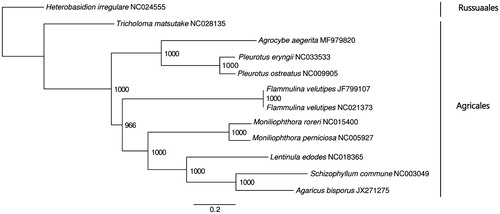
The complete mitogenome of A. aegerita was 116,329 bp in length, containing two pairs of large reverse repeat regions (4061 bp and 24,473 bp in length, respectively). The overall GC content was 27.6%, including 17 protein-coding genes, two ribosomal protein gene rps3, 32 tRNA genes, and two rRNA genes (rnl and rns). The protein coding genes involved in respiration and oxidative phosphorylation included the three ATP synthase subunits (atp6, atp8 and atp9), the seven NADH dehydrogenase subunits (nad1, nad2, nad3, nad4, nad4L, nad5 and nad6), the three cytochrome oxidase subunits (cox1, cox2 and cox3), and the apocytochrome b (cob). The 32 tRNA genes ranged in size from 71 bp to 87 bp, and coded for all 20 standard amino acids. The ML tree showed that A. aegerita is sister to the genus Pleurotus forming a separated clade from other Agaricales species (). Support was high for all nodes, but the close relationship between Agrocybe and Pleurotus differs from the previous studies (Matheny et al. Citation2006), suggesting that adding more complete mitogenome will greatly help to retrieve a high resolution and highly supported phylogeny. With this first complete mitochondrial genome of Agrocybe aegerita, we expect identification in Agrocybe will be facilitated, and breeding programs improved and expanded.
Acknowledgements
The authors would like to acknowledge the technical staff of the Biology Institute, Guangxi Academy of Sciences for their assistance in access to the A. aegerita strain SWS_17.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Barroso G, Blesa S, Labarère J. 1995. Wide distribution of mitochondrial genome rearrangements in wild strains of the cultivated basidiomycete Agrocybe aegerita. Appl Environ Microbiol. 61:1187–1193.
- Chen WM, Chai HM, Zhou HM, Tian GT, Li SH, Zhao YC. 2012. Phylogenetic analysis of the Agrocybe aegerita multispecies complex in Southwest China inferred from ITS and mtSSU rDNA sequences and mating tests. Ann Microbiol. 62:1791–1801.
- Hinsinger DD, Strijk JS. 2016. Toward phylogenomics of Lauraceae: the complete chloroplast genome sequence of Litsea glutinosa (Lauraceae), an invasive tree species on Indian and Pacific Ocean islands. Plant Gene. 9:71–79.
- Jiang GF, Hinsinger DD, Strijk JS. 2016. Comparison of intraspecific, interspecific and intergeneric chloroplast diversity in Cycads. Sci Rep. 6:31473.
- Matheny PB, Curtis JM, Hofstetter V, Aime MC, Moncalvo JM, Ge ZW, Slot JC, Ammirati JF, Baroni TJ, Bougher NL, et al. 2006. Major clades of Agaricales: a multilocus phylogenetic overview. Mycologia. 98:982–995.
- Noël T, Labarère J. 1994. Homologous transformation of the edible basidiomycete Agrocybe aegerita with the URA1 gene: characterization of integrative events and of rearranged free plasmids in transformants. Curr Genet. 25:432–437.
- Posada D. 2009. Selection of models of DNA evolution with jModelTest. Methods Mol Biol. 537:93–112.
- Singer R. 1986. The Agaricales in modern taxonomy. Koenigstein (Germany): Koeltz Scientific Books.
- Thushara D, Vanisree M, Gary M, Davidl DW, Muraleedharang N. 2008. Health-beneficial qualities of the edible mushroom, Agrocybe aegerita. Food Chem. 108:97–102.
- Tsai SY, Huang SJ, Mau JL. 2006. Antioxidant properties of hot water extracts from Agrocybe cylindracea. Food Chem. 98:670–677.
- Uhart M, Sirand-Pugnet P, Labarère J. 2007. Evolution of mitochondrial SSU-rDNA variable domain sequences and rRNA secondary structures, and phylogeny of the Agrocybe aegerita multispecies complex. Res Microbiol. 158:203–212.
- Wasser SP, Weis AL. 1999. Therapeutic effects of substances occurring in higher Basidiomycetes mushrooms: a modern perspective. Crit Rev Immunol. 19:65–96.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhao CG, Sun H, Tong X, Qi YP. 2003. An antitumour lectin from the edible mushroom Agrocybe aegerita. Biochem J. 374:321–327.
