Abstract
Nicotiana attenuata (N. attenuata) is a species of wild tobacco, which is a predominant ecological model native to North America. Here, we first assembled the complete mitochondrial (mt) DNA sequence of N. attenuata into a circular genome of length 394,341 bp, including 40 protein-coding genes, 4 rRNA genes, and 23 tRNA genes. A neighbour-joining phylogenetic tree was constructed based on the complete mt genome of N. attenuata and other 27 plant species to assess their phylogenic relationship and evolution. The complete mt genome of N. attenuata would be useful for the further analyses of Nicotiana species.
Nicotiana attenuata, popularly known as coyote tobacco, is an annual herb that is native to western North America. Thanks largely to its diverse interactions with plenty of different plants, insects, and microorganisms in its native habitat, N. attenuata has been utilized as an ecological model species since 1994 (Baldwin et al. Citation1994). Additionally, N. attenuata is a species that can induce nicotine, volatiles and phenolics in response to damage (Dam et al. Citation2001).
Mitochondria encode the main manufactures of cellular ATP, and play vital roles in the regulation of cellular metabolism (Lei et al. Citation2013; Bi et al. Citation2016). In plant species, the mitochondrial (mt) genomes vary considerably in length, gene content and order (Richardson et al. Citation2013). Until now, there are only 274 complete plant mt genomes deposited in GenBank Organelle Genome Resources. In this study, we assembled the complete mt genome sequence of N. attenuata, which may provide more desirable information for better understanding the interactions of N. attenuata in its native environment and the nicotine biosynthetic pathway in Nicotiana species.
Sample of N. attenuata used for extraction was harvested from a native population in Washington Country, Utah (Geographic coordinate: 37° 16′ 48″ N, 113° 31′ 12″ W). The genomic DNA was isolated from late rosette-stage plants following the CTAB DNA extraction protocol (Bubner et al. Citation2004), and then deposited in Max Planck Institute for Chemical Ecology (Jena, Germany). In this study, the complete mt DNA sequence of N. attenuata was assembled into a gap-free and accurate genome of length 394,341 bp by Newbler v3.0 and MacVector v15.1.3, and then submitted to GenBank with the accession number MF579563. The GC content of the mt genome is 45.05%, a widespread value in most Asterids.
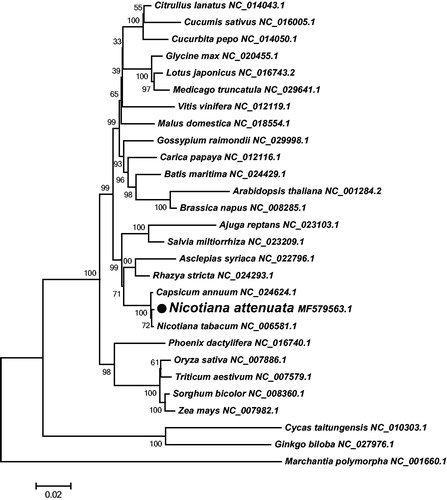
Using the annotation program MITOFY (Alverson et al. Citation2010), a total of 67 genes were identified in the mt genome of N. attenuata, including 40 protein-coding genes, 23 tRNA genes, and four rRNA genes. Among these, eight genes contain two copies (rpl5, rps14, rrn26, trnM-CAT, trnfM-CAT, trnN-GTT, trnP-TGG, and trnE-TTC). Additionally, 33 exons and 14 introns were identified in nine protein-coding genes (rps3, cox2, nad1, nad2, nad5, nad7, rps10, ccmFc, and rpl2). The analysis of codon usage shows that most of the mt protein-coding genes share the common start codon: ATG, except rps10, cox1 and rps4, which use ACG (C to U RNA-editing) as start codon. Three types of stop codons are found in the mt protein-coding genes: TAA (20 genes: atp1, atp8, cox1, cox2, nad1, nad2, nad3, nad4L, nad5, nad6, nad9, rpl2, rpl5x2, rpl10, rpl16, rps1, rps4, rps19, and sdh4), TGA (13 genes: atp6, ccmB, ccmC, ccmFc, ccmFn, cob, cox3, matR, nad4, rps10, rps12, rps13, and sdh3), and TAG (seven genes: atp4, atp9, mttB, nad7, rps3, and rps14x2). A neighbour-joining phylogenetic tree was constructed using 28 plant mt genomes based on 23 conserved protein-coding genes, including 18 respiratory complex genes (atp1, atp4, atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, and nad9), four cytochrome c biogenesis genes (ccmB, ccmC, ccmFc, and ccmFn) and one Maturase gene (matR). As shown in , the phylogenetic tree exhibited that N. attenuata and Nicotiana tabacum are classified into one clade, and the clade of Nicotiana was evolutionarily close to the Solanaceae plant Capsicum annuum.
Acknowledgements
We would like to acknowledge the Max Planck Institute for Chemical Ecology for their assistance in sampling the material of Nicotiana attenuata.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Alverson AJ, Wei XX, Rice DW, Stern DB, Barry K, Palmer JD. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 27:1436.
- Baldwin IT, Staszak-Kozinski L, Davidson R. 1994. Up in smoke: I. Smoke-derived germination cues for postfire annual, Nicotiana attenuata torr. Ex. Watson. J Chem Ecol. 20:2345.
- Bi C, Paterson AH, Wang X, Xu Y, Wu D, Qu Y, Jiang A, Ye Q, Ye N. 2016. Analysis of the complete mitochondrial genome sequence of the diploid cotton Gossypium raimondii by comparative genomics approaches. BioMed Res Int. 2016:5040598.
- Bubner B, Gase K, Baldwin IT. 2004. Two-fold differences are the detection limit for determining transgene copy numbers in plants by real-time PCR. BMC Biotechnol. 4:14.
- Dam NMV, Horn M, Mareš M, Baldwin IT. 2001. Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J Chem Ecol. 27:547–568.
- Lei B, Li S, Liu G, Chen Z, Su A, Li P, Li Z, Hua J. 2013. Evolution of mitochondrial gene content: loss of genes, tRNAs and introns between Gossypium harknessii and other plants. Plant Syst Evol. 299:1889–1897.
- Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. 2013. The “fossilized” mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 11:29.