Abstract
The complete mitochondrial DNA sequence of the South America fur seal (Arctocephalus australis) was obtained by a shotgun sequencing approach. The mitogenome is 16,372 bp in length and includes the genes coding for the two rRNA species (12S and 16S), 13 protein-coding genes, 22 transfer RNA genes, and a control region. The base composition is 33.0% for A, 26.7% for C, 26.1 for T and 14.2% for G, with an overall GC content of 40.9%. The description of this mitogenome will be useful for further phylogeny and genetic studies on Pinnipeds.
The South American fur seal (Arctocephalus australis) is a Neotropical otariid species that occurs on the western South Atlantic and the eastern South Pacific coasts of South America (King Citation1983; Jefferson et al. Citation2015). Despite the intensive hunt for centuries, especially during the eighteenth and nineteenth centuries, leaving the species at the edge of extinction (Bonner Citation1982), the species is considered as Least Concern by the IUCN based on the thriving of the species in the last decades in the Atlantic Ocean (Cárdenas-Alayza et al. Citation2016). However, there are concerns on the conservation status of subpopulations in Peru and Southern Chile (Cárdenas-Alayza et al. Citation2016).
The use of genetic approaches is vital to understand the historical events that lead to the present distribution of species. The genomic mapping meaningfully increases the knowledge on species genome organization, allowing the genetic screen, identification, and location of major genes on populations (Crittenden et al. Citation1993), which could be used on the development of management and conservation measures, especially on species distributed along wide areas.
Total genomic DNA was extracted from liver tissue of five A. australis collected from necropsies performed at Isla de Guafo (43°35′ S, 74°42′ W), Chile, during Austral summer of 2016 and 2017, using a commercial kit (E.Z.N.A.®, Omega bio-tek, GA) and later sequenced on a MiSeq®, Illumina (San Diego, CA). The specimens were deposited on the Instituto de Patología Animal, Facultad de Ciencias Veterinarias, Universidad Austral de Chile (IPAFCV/N2/2016, IPAFCV/N6/2016, IPAFCV/N10/2016, IPAFCV/N4/2017, and IPAFCV/N5/2017). Raw reads were trimmed for Nextera DNA Library Preparation Kit (Illumina, CA) adaptors with Trimmomatic (Bolger et al. Citation2014) and by quality with Prinseq (Schmieder and Edwards Citation2011), and filtered for high-quality sequences using the public Galaxy server (http://galaxyproject.org). Reads were assembled using NextGENe® software (Softgenetics®, State College, PA). A local BLAST search was used to select reads containing mitochondrial genome. The mitochondrial genomes of the A. townsendi (NC008420) and A. forsteri (KT693374) were downloaded and used to identify contigs containing mitochondrial sequences. The complete mitogenome was assembled and annotated in Geneious 9.1.2 (Kearse et al. Citation2012). CLUSTALW alignments against reference mitochondrial genomes were used to confirm manual annotations.
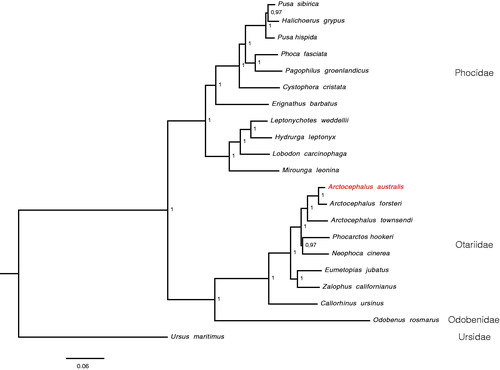
The mitochondrial genome of the A. australis (Accession MG023139) is 16,372 bp long, and includes 13 protein-coding genes, 22 transfer RNA genes, and a control region. The base composition of the mitogenome is 33.0% for A, 26.7% for C, 26.1 for T, and 14.2% for G, with an overall GC content of 40.9%. The overall mitogenome is 98% similar to A. forsteri and 95% to A. townsendi. The reconstructed phylogeny supported the placement of A. australis as sister to other Arctocephalus clade (). Here, we report for the first time the complete mitochondrial genome of the A. australis, which could be important for further phylogenetic and population genetic studies on pinnipeds, especially of the genus Arctocephalus.
Acknowledgements
This work was performed under authorization of the Subsecretaria de Pesca y Agricultura de Chile (SUBPESCA), permit number “Resolución exenta No. 88 del 15 de Enero del 2014”. The authors appreciate the help in the field of Francisco Muñoz and the technical and logistic support of the Chilean Navy Guafo Island lighthouse personnel.
Disclosure statement
All the experiments comply with the current laws of the Republic of Chile and the authors declare that they have no conflict of interest.
Additional information
Funding
References
- Bonner WN. 1982. Seals and man – a study of interactions. Seatle, CA: University of Washington Press.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Cárdenas-Alayza S, Oliveira L, Crespo E. (2016). Arctocephalus australis. The IUCN Red List of Threatened Species 2016: e.T2055A45223529; [accessed 2014 Sep 04]. Available from: http://dx.doi.org/10.2305/IUCN.UK.2016-1.RLTS.T2055A45223529.en.
- Crittenden LB, Provencher L, Levin I, Abplanalp H, Briles RW, Dodgson JB. 1993. Characterization of a red jungle fowl by white leghorn backcross reference population for molecular mapping of the chicken genome. Poultry Sci. 72:334–348.
- Jefferson TA, Webber MA, Pitman RL, Gorter U. (2015). Marine mammals of the world: a comprehensive guide to their identification. 2nd ed. Waltham, MA, USA: Academic Press.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- King JE. 1983. Seals of the world. 2nd ed. NY: Cornell University Press.
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 27:863–864.