Abstract
The mitogenome sequence of Sponge Halichondria okadai (Kadota, 1922) (Suberitida, Halichondriidae) was determined for the first time in this study. The circular genome is 20,722 bp in length, containing 14 protein coding genes (PCGs), two ribosomal RNAs (rRNAs), and 25 transfer RNAs (tRNAs). The nucleotide composition of mitogenome consists of 29.5% A, 14.2% C, 21.5% G, 34.7% T, showing a high content of A + T similar to the other Suberitid sponges. These results will be useful for inferring the phylogenetic relationships among the members of family Halichondriidae within the Suberitids.
Halichondria okadai is a thin encrusting species belonging to family Halichondriidae and inhabits in the intertidal zone. Sponges of the genus Halichondria Fleming, 1828 are presently recognized as 93 valid species in the world ocean (van Soest et al. Citation2017) and they are distributed over all regions and habitats (Hooper and van Soest Citation2002). Twelve species of the genus Halicondria have been reported from Korean waters (NIBR Citation2012). It is known that the drug Eribulin extracted from H. okadai has the effect of extending the life span of women with breast cancer (Cortes et al. Citation2011; Ohno et al. Citation2011). Until now, the only two species (Halichondria sp., Hymeniacidon sinapium) of Halichondriid have been reported for their complete mitochondrial genomes (Jun et al. Citation2015; Wang et al. Citation2016). Here we report the complete mitochondrial genome (mitogenome) of H. okadai for the first time.
Specimens of H. okadai were collected from intertidal zone of Jeju Island of Korea. The voucher specimens were deposited in National Marine Biodiversity Institute of Korea (MABIK IV00163014-00163019). The genomic DNA was isolated from the tissue and the mitogenome sequences were analysed by application of Illumina Hiseq2000 sequencing platform (Macrogen, Seoul, Korea). The sequences were assembled and annotated in comparison with the previously reported mitogenome sequences of Halichondriid species (Jun et al. Citation2015; Wang et al. Citation2016) using Geneious v9.1.8 (Kearse et al. Citation2012). Phylogenetic tree was constructed using MEGA6 (Tamura et al. Citation2013).
The complete mitogenome of Halichondria okadai (GeneBank accession number MG267395) is 20,722 bp in length, containing 14 protein coding genes (PCGs), two ribosomal RNAs (rRNAs), and 25 transfer RNAs (tRNAs). The overall nucleotide base composition of H. okadai is 29.5% A, 14.2% C, 21.5% G, 34.7% T, showing a high A + T bias (64.2%) similar to the other Halichondriid sponges. All PCGs use the typical ATG as the start codon. Ten PCGs (atp9, cytb, cox3, atp6, cox2, nad1, cox1, nad4l, nad6, and nad4) use TAA as the stop codon while four (atp8, nad5, nad2, nad3) genes have TAG. The lengths of tRNA genes range from 71 to 80 bp and all tRNAs have the typical clover leaf structure. The sizes of 12S and 16S rRNAs are 1364 bp and 2834 bp, respectively.
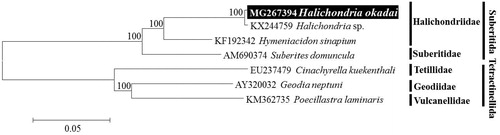
The taxonomic position of H. okadai analysed by molecular phylogenetic tree of combined 14 PCGs sequences using the neighbour-joining (NJ) method with the K2P model in MEGA 6. H. okadai were clustered together as Halichondria sp. (KX244759), previously announced from GenBank. Their cluster is strongly supported by bootstrap values of 100% (). The similarity of mitogenome between H. okadai and Halichondria sp. is 99.6%, they were same species. To conclude, we confirmed that Halichondria sp. reported by Wang et al. (Citation2016) was H. okadai. In this study, we determined the complete mitogenome of sponge H. okadai, and it will provide useful information for understanding of evolutionary history and phylogeny of the genus Halichondria in relation to the other genera within family Halichondriidae.
Figure 1. Neighbour-joining (NJ) tree based on the mitogenome sequences of three Halichondriid species including Halichondria okadai with two other related species in Suberitida. Three species (Cinachyrella kuekenthali, Geodia neptuni, Poecillastra laminaris) derived from Tetractinellida was used as outgroup for tree rooting. Numbers above the branches indicate NJ bootstrap values from 1000 replications.
Disclosure statement
The authors declare that they do not have any conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Cortes J, O'Shaughnessy J, Loesch D, Blum JL, Vahdat LT, Petrakova K, Chollet P, Manikas A, Diéras V, Delozier T, et al. 2011. Eribulin monotherapy versus treatment of physician's choice in patients with metastatic breast cancer (EMBRACE): a phase 3 open-label randomised study. Lancet. 377:914–923.
- Hooper JNA, Van Soest RWM. 2002. Systema Porifera: a guide to the classification of sponges. New York (UK): Kluwer Academics/Plenum Publisher Press; p. 1–1101.
- Jun J, Yu JN, Choi EH. 2015. Complete mitochondrial genome of Hymeniacidon sinapium (Demospongiae, Halichondriidae). Mitochondrial DNA. 26:261–262.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- National Institute of Biological Resources (NIBR). 2012. National list of species of Korea “Invertebrates-I”. National Institute of Biological Resources; p. 26–27.
- Ohno O, Chiba T, Todoroki S, Yoshimura H, Maru N, Maekawa K, Imagawa H, Yamada K, Wakamiya A, Suenaga K, et al. 2011. Halichonines A, B, and C, novel sesquiterpene alkaloids from the marine sponge Halichondria okadai Kadota. Chem Commun (Camb). 47:12453–12455.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Van Soest RWM, Boury-Esnault N, Hooper JNA, Rützler K, de Voogd NJ, Alvarez de Glasby B, Hajdu E, Pisera AB, Manconi R, Schoenberg C, et al. 2017. World Porifera Database; [cited 2017 Sep 28]. http://www.marinespecies.org/porifera.
- Wang D, Zhang Y, Huang D. 2016. The complete mitochondrial genome of sponge Halichondria (Halichondria) sp. (Demospongiae, Suberitida, Halichondriidae). Mitochondrial DNA B. 1:512–514.
