Abstract
Pyrhila pisum is known as leucosiid crab. So far, there is no study about whole mitochondrial genome in Leucosiidae family. Here, we report first the complete sequence of the mitochondrial genome from P. pisum, which is composed of 15,516 base pair encoding 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs, and A + T-rich region. The nucleotide composition of P. pisum was G + C: 25.5%, A + T: 74.5%, with a strong AT bias. In phylogenetic analysis using whole mitogenome, it was figured out that P. pisum was closely related to Sesarma neglectum but their family is different.
Keywords:
Pyrhila pisum is classified in Leucosiidae family according to morphological classification (Galil Citation2009). This crab mostly inhabits tidal flats, continental coastal zone, and subtropical coasts in Asia such as China, Japan, Taiwan, and Korea. The preferences of feeds are changed by time and sex but usually it prefers bivalves especially Arcuatula senhousia and Venerupis philippinarum (Kobayashi Citation2013). At present, this species is considered a threatened endangered species due to reduction of tidal flats (Kobayashi and Archdale Citation2017). In this study, we sequenced the mitochondrial genome of P. pisum and figured out phylogenetic relationship to other organisms through comparative genomics.
A sample of P. pisum was collected in Gangseo-gu, Busan, Korea (GIS: 35°04′47″N 128°52′36″E). The whole body specimen is registered under the voucher number NIBRGR0000099840 in National Institute of Biological Resources, Korea. The mitochondria were separated from 1 g of muscle tissue by differential centrifugation (Graham Citation2001). The mitochondrial DNA was extracted using QIAamp Tissue Kit (Qiagen, Valencia, CA). To amplify the mitogenome, specific PCR primers for Decapoda were used (Martin et al. Citation2016). Additionally, PCR primers based on conserved 16S rRNA gene and cox1 were designed and tried as well. PCR temperature for extension was gradually decreased than regular cycle in order to pcrevent PCR inhibition caused by A + T-rich region (Su et al. Citation1996). After purification of PCR amplicons, NGS library preparation and sequencing were performed using the Ion Torrent PGM platform following the manufacturer’s instructions (Life Technologies/Thermo Fisher Scientific, Waltham, MA). Whole mitochondrial genome assembly and annotation were proceeded using the CLC genomics workbench 8.5 (CLC Bio, Aarhus, Denmark). Protein-coding gene (PCGs), transfer RNA (tRNAs), and ribosomal RNA (rRNAs) were confirmed by NCBI Basic Local Alignment Search Tool (BLAST) and tRNA-scan 1.21 (Altschul et al. Citation1990; Lowe and Eddy Citation1997).
The assembled mitogenome was composed of circular DNA with 15,516 bp containing 13 protein-coding gene (PCGs), two ribosomal RNA (rRNAs), 22 transfer RNA (tRNAs) and A + T-rich region. In 13 PCGs, three kinds of start codon such as ATG, ATA and ATT were found. Most of PCGs contained TAA stop codon except cox1, cox2, cox3, and cytb. These four PCGs were terminated by incomplete stop codon such as T(aa) or TA(a) (Yamauchi et al. Citation2003; Hou et al. Citation2007; Zhang et al. Citation2016). The overall nucleotide composition was asymmetric (A: 37.28%, T: 37.20%, G: 10.22%, C: 15.30%) with a strong AT bias. The complete mitogenome of P. pisum was submitted to GenBank (accession no. NC030047).
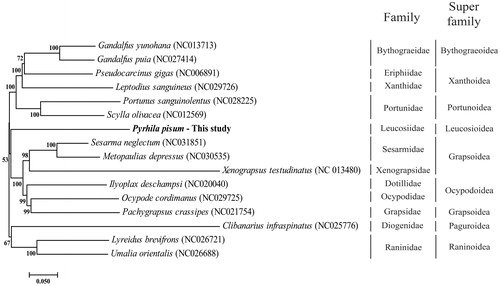
The Phylogenetic tree of P. pisum and other crabs in Decapoda order was conducted using MEGA 6.0 by neighbour-joining method with 1000 replicate bootstrap (Tamura et al. Citation2013). The phylogenetic location of P. pisum was close to Grapsoidea super family (). Since there are limited mitogenome information in family of Leucosiidae, more sequence reports are needed to classify of Decapoda order. This result emphasize that the complete mitogenome sequence of P. pisum can contribute to the phylogenetic knowledge of the genus Pyrhila as well as family Leucosiidae.
Figure 1. Phylogenetic tree of P. pisum with other crab in Decapoda order. The number in phylogenetic tree is bootstrap value. On the right side, vertical stick indicated specific family and super family of crab in Decapoda order. The GenBank accession numbers are indicated after the scientific name.
Disclosure statement
No potential conflict of interest was reported by the authors.
Reference
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Galil BS. 2009. An examination of the genus Philyra Leach, 1817 (Crustacea, Decapoda, Leucosiidae) with descriptions of seven new genera and six new species. Zoosystema. 31:279–320.
- Graham JM. 2001. Current protocols in cell biology: isolation of mitochondria from tissues and cells by differential centrifugation, Vol. 3 . Liverpool: John Wiley & Sons, Inc.; p. 1–15.
- Hou W, Chen Y, Wu X, Hu J, Peng Z, Yang J, Tang Z, Zhou C-Q, Li Y-m, Yang S. 2007. A complete mitochondrial genome sequence of Asian black bear Sichuan subspecies (Ursus thibetanus mupinensis). Int J Biol Sci. 3:85–90.
- KOBAYASHI S. 2013. Feeding habits of the Leucosiid Crab Pyrhila pisum (De Haan) observed on a sandy tidal flat in Hakata bay, Fukuoka, Japan. Jpn J Benthol. 68:37–41.
- Kobayashi S, Archdale MV. 2017. Occurrence pattern and reproductive ecology of the leucosiid crab Pyrhila pisum (De Haan) in tidal flats in Hakata Bay, Fukuoka, Japan. Crustacean Res. 46:103–119.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Martin JW, Crandall KA, Felder DL. 2016. Decapod crustacean phylogenetics. New York (NY). CRC Press.
- Su X-z, Wu Y, Sifri CD, Wellems TE. 1996. Reduced extension temperatures required for PCR amplification of extremely A + T-rich DNA. Nucleic Acids Res. 24:1574–1575.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Yamauchi MM, Miya MU, Nishida M. 2003. Complete mitochondrial DNA sequence of the swimming crab, Portunus trituberculatus (Crustacea: Decapoda: Brachyura). Gene. 311:129–135.
- Zhang Q-X, Guan D-L, Niu Y, Sang L-Q, Zhang X-X, Xu S-Q. 2016. Characterization of the complete mitochondrial genome of the Asian planthopper Ricania speculum (Hemiptera: Fulgoroidea: Ricannidae). Conservation Genetics Resourc. 8:463–466.
