Abstract
We sequenced the complete mitochondrial genomes of three pairs of congeneric peripheral fishes distributed on either side of the Isthmus of Panama in order to test their status as geminate species pairs. Our phylogenetic analysis did not support a sister relationship between Gobiomorus dormitor and G. maculatus and therefore they cannot be considered geminates. The average genetic distance of protein-coding genes between Sicydium altum and S. salvini was more than two times larger than between Atlantic and Pacific Awaous banana, suggesting different timings for their divergence across the Isthmus of Panama.
Introduction
The formation of the Isthmus of Panama represents one of the most dramatic geological events in Earth’s history with major climatic and biological implications. Despite its global importance, the timing of the uplift of the Isthmus of Panama remains an open question that has resulted in a thrilling scientific discussion (Montes et al. Citation2012; Bacon et al. Citation2015a, Citation2015b; Lessios Citation2015; O’Dea et al. Citation2016; Jaramillo et al. Citation2017), in which the most recent studies challenge the long-held idea of a Pliocene connection (ca. 3.5 Million years ago) of North and South America (Coates and Obando Citation1996). Thus, recent works propose an earlier age (i.e. Miocene-Oligocene) for the formation of the Isthmus, as well as a more complex geological history than previously believed (Montes et al. Citation2012).
The formation of the Isthmus created a landbridge that allowed species to cross between South and North America, and, at the same time, created a barrier to gene flow between marine organisms in the Pacific and Caribbean. Provided this barrier, species may have diverged on either side of the Isthmus into descendant sister taxa called ‘geminate species’ (Jordan Citation1908). Molecular evidence indicates that all geminate species were not simultaneously separated by the emerging Isthmus (Knowlton and Weigt Citation1998; Marko Citation2002; Lessios Citation2008; Miura et al. Citation2010; O’Dea et al. Citation2016), and suggests that ecological differences, such as habitat preference, may influence the timing of separation. For example, mangrove dwelling invertebrates exhibit fewer genetic differences than species associated with deeper habitats (Knowlton and Weigt Citation1998; Miura et al. Citation2010). Similarly, peripheral fishes (i.e. groups with a recent marine origin that have invaded coastal streams; Myers Citation1949) of the genus Dormitator show remarkably young divergence times (Galván-Quesada et al. Citation2016). These results support the idea that near-shore taxa are ideal candidates for molecular clock calibrations, because their limited genetic divergences might reflect the latter stages of transisthmian marine connection (Knowlton and Weigt Citation1998; Galván-Quesada et al. Citation2016).
In this study, we provide a comparison of genetic distances across complete mitochondrial genomes of three pairs of peripheral fish species, or lineages, with distributions on either side of the Isthmus of Panama. We infer their evolutionary relationships to test the hypothesis that they are geminate species.
Materials and methods
We collected specimens from six species or lineages of gobiiform fishes distributed in Atlantic and Pacific watersheds of Panama: Gobiomorus dormitor (STRI-15207, Río Chagres, Río Chagres, Panama Province, Panama), Gobiomorus maculatus (STRI-7077, Río Pavo, Río Pavo, Veraguas Province, Panama), Sicydium altum (STRI-4981, Río Bongie, Río Changuinola, Bocas del Toro Province, Panama), S. salvini (STRI-660, Río Chiriquí Viejo, Río Chiriquí Viejo, Chiriquí Province, Panama; STRI-3170, Río Lajas, Río Santa María, Herrera Province, Panama), Awaous banana (STRI-4986, Río Bongie, Río Changuinola, Bocas del Toro Province, Panama), and A. banana (STRI-11209, Río Mamoní, Río Bayano, Panama Province, Panama). We excised the gill arches and preserved them in a 20% dimethyl sulphoxide (DMSO), 0.5M EDTA, pH 8 solution at 4 °C until DNA extraction. All specimens were initially fixed in formalin, later transferred to 70% ethanol, and deposited and vouchered in the Neotropical Fish Collection (NFC-STRI) at the Smithsonian Tropical Research Institute in Panama (STRI).
We recovered complete mitochondrial DNA sequences as a by-product of hybrid target capture of ultraconserved elements (UCEs) (Faircloth et al. Citation2012). Briefly, we extracted DNA using a QIAGEN DNeasy kit (Qiagen, Valencia, CA) and randomly sheared 400–1000 ng of DNA by sonication to a target size of 400–600 base pairs (bp). We constructed genomic libraries for each of our samples using the Kapa Hyper Prep Kit v.3.15 (Kapa Biosystems, Wilmington, MA), and enriched them for 500 UCE loci using the Actinops-UCE-0.5Kv1 probe set (Faircloth et al. Citation2013) following protocols for the MYcroarray MYBaits kit v.3.0 (MYcroarray, Ann Arbor, MI). Then we sequenced these libraries in one lane of PE150 Illumina MiSeq (Illumina, San Diego, CA).
We trimmed raw reads for adapter contamination and low quality bases in illumiprocessor (Faircloth Citation2013), and mapped them to reference mitogenomes using Bowtie 2 (Langmead and Salzberg Citation2012) as implemented in Geneious 10.1.3 (www.geneious.com, Kearse et al. Citation2012). Also using Geneious 10.1.3, we created and annotated consensus sequences.
We aligned our mitogenomes with representative mitogenomes from 22 species belonging to five out of the seven families of Gobiiformes (sensu Agorreta et al. Citation2013), including three newly sequenced mitogenomes of Neotropical eleotrids (Alda et al. Citation2017), and Kurtus gulliveri (Kurtiformes) that was used as an outgroup. We extracted and individually re-aligned all 13 protein-coding genes for subsequent phylogenetic analysis. We partitioned the data by gene and by codon, and estimated the best partition scheme under the GTR + GAMMA substitution model using PartitionFinder (Lanfear et al. Citation2012). We used these partitions in a Maximum Likelihood analysis in RAxML (Stamatakis Citation2014), in which we ran 40 searches to find the best tree, and performed 500 bootstrap replicates to assess nodal support.
To compare mitochondrial divergences across pairs of putative geminate taxa, we calculated uncorrected p-distances between congeneric taxon pairs for each mitochondrial gene, and calculated their standard errors using 500 bootstrap replicates in MEGA7 (Kumar et al. Citation2016).
Results and discussion
We obtained complete mitochondrial genome sequences with a minimum of 10× coverage for three species pairs, or lineages, of peripheral gobiiform fishes: G. dormitor (MF927493) and G. maculatus (MF927494); S. altum (MF927496) and S. salvini (MF927497, MF927498); and A. banana from Atlantic (MF927488) and Pacific (MF927489) watersheds.
The size of the mitochondrial genomes ranged from 16,474 bp in G. dormitor to 16,536 bp in G. maculatus. All species contained 22 tRNA genes, two rRNA genes, 13 protein coding regions, as well as control region arranged in the same order. The most common start codon ATG was found in all genes except in COXI that had the start codon GTG, and in ND6 that had TAC as the start codon. The stop codon TAA terminated the genes ND2, ND4L and ATP8 in all species, whereas genes ND1 and ND5 exhibited either TAA or TAG as stop codons. Incomplete stop codons were found in some or all species for genes ATP6, COXII, COXIII, CYTB, ND3 and ND4 formed of only T or TA, which are likely completed as TAA via post-transcriptional polyadenylation.
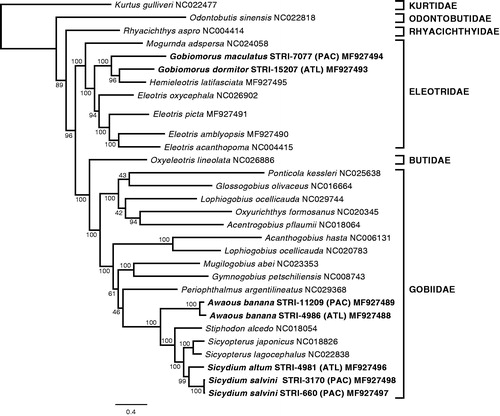
Our phylogenetic analysis was congruent in the relationships among families and genera with recent studies using both mitochondrial and nuclear loci (Agorreta et al. Citation2013; Adrian-Kalchhauser et al. Citation2017). The species or lineage pairs of Sicydium and Awaous showed sister relationships and can be considered as geminate species. However, G. dormitor and G. maculatus were not recovered as sister species, and consequently the hypothesis that they are geminate species was rejected (). This result is in contrast with the results of Thacker and Hardman (Citation2005) and Thacker (Citation2017), who analyzed the same data set in a phylogeny of the Gobiodei and in a comparative morphological analysis of geminate species, using Maximum Parsimony and Bayesian Inference, respectively. In both cases they recovered a sister relationship for the two species of Gobiomorus. Conversely, further reanalyses of the data from Thacker and Hardman (Citation2005) have also evidenced the non-monophyly of Gobiomorus, and similarly to our study, have recovered a sister relationship between G. dormitor and H. latifasciata (Thacker Citation2009; Agorreta and Rüber Citation2012). Furthermore, and in agreement with the latter, a recent barcoding study with a broader geographical and taxonomical sampling within the Eleotridae, pointed to the Eastern Pacific species G. polylepis as the sister of G. dormitor, and H. latifasciata as sister to the two of them (Guimarães-Costa et al. Citation2017). According to this, G. dormitor and G. polylepis would constitute the geminate species pair in the genus Gobiomorus.
Figure 1. Maximum-likelihood (RAxML) phylogeny using 13 protein-coding mitochondrial genes from a selection of species of Gobiiformes (GenBank accession numbers indicated), and Kurtus gulliveri, which was used as the outgroup. Numbers next to nodes are support values obtained after 500 bootstrap replicates. Sequences from specimens obtained in this study are highlighted in bold.
As would be expected, given they were not recovered as sister groups, G. dormitor and G. maculatus exhibited the largest divergences for every mitochondrial gene among the species compared (). Distances were as low as 1.37% for tRNA Leu, and as large as 31.14% for ND6. On average, protein-coding genes were 18.74% ± 4.70 divergent. These genetic distances were also greater than between G. dormitor and H. latifasciata (mean = 15.43% ± 1.82), except for the COXII gene (12.74% versus 12.30%).
Figure 2. Genetic divergence across all mitochondrial genes between three pairs of species: Gobiomorus dormitor versus G. maculatus, Awaous banana from Atlantic versus A. banana from the Pacific watersheds, and Sicydium salvini versus S. altum. Genes are presented in the same order as they appear in these species mitochondrial genomes. Error bars represent standard errors and the dashed horizontal line indicates the commonly used threshold at 2% genetic divergence.
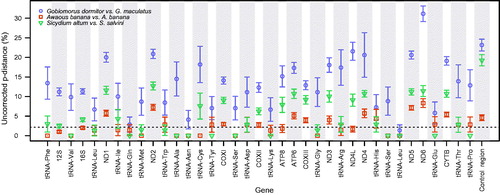
Throughout all comparisons, control region and ND6 were two of the most variable genes, while tRNA genes were consistently the least variable. The fewest differences existed between Atlantic and Pacific A. banana: ten tRNA genes were identical, with the remaining 12 ranging from 1.33 to 4.34% different. All protein-coding genes were over 2% divergent (mean = 4.75% ± 2.02), which is a common threshold for species delimitation in mitochondrial and barcoding studies (Ward Citation2009; Pereira et al. Citation2013; Bagley et al. Citation2015), except for ATP8 and ND4L (1.82% and 1.68% divergent, respectively). Sicydium altum and S. salvini divergences ranged between 1.66 and 12.70% for the protein coding genes (mean = 10.03% ± 1.66) ().
The divergence values between Awaous and Sicydium geminate pairs are in the lower and upper boundaries of other geminate species pairs of peripheral fishes (e.g. D. maculatus versus D. latifrons: CYTB uncorrected p-distance = 8.6%, Galván-Quesada et al. Citation2016; and Caribbean Agonostomus monticola versus Pacific A. monticola: CYTB uncorrected p-distance = 8.3%, McMahan et al. Citation2013). Assuming that mutation rates are similar across evolutionary close species, these differences might reflect a range of divergence times across geminate species of peripheral fishes that could be related to their habitat preferences. For example, Sicydium species inhabit fast flowing rivers and creeks between sea level and 1100 m, whereas A. banana is commonly found in estuaries and rivers only up to 300 m above sea level. Therefore, the affinity of Awaous to near-shore environments and brackish waters may have allowed them to maintain gene flow until the very last instances of marine connectivity across the Central American Seaway. Overall, these new mitogenomes constitute an important resource for future investigations on rates of molecular evolution and divergence times of geminate species, and eventually for a further understanding of the geological history and age of the Isthmus of Panama.
Acknowledgements
The authors thank Ruth G. Reina for laboratory and curatorial assistance. The authors also acknowledge Brant Faircloth for leading the workshop during which these data were generated.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Adrian-Kalchhauser I, Svenss O, Kutschera VE, Rosenblad MA, Pippel M, Winkler S, Schloissnig S, Blomberg A, Burkhardt-Holm P. 2017. The mitochondrial genome sequences of the round goby and the sand goby reveal patterns of recent evolution in gobiid fish. BMC Genomics. 18:177.
- Agorreta A, Rüber L. 2012. A standardized reanalysis of molecular phylogenetic hypotheses of Gobioidei. Syst Biodivers. 10:375–390.
- Agorreta A, San Mauro D, Schliewen U, Van Tassell JL, Kovacic M, Zardoya R, Rüber L. 2013. Molecular phylogenetics of Gobioidei and phylogenetic placement of European gobies. Mol Phylogenet Evo. 69:619–633.
- Alda F, Adams AJ, McMillan WO, Chakrabarty P. 2017. Complete mitochondrial genomes of three Neotropical sleeper gobies: Eleotris amblyopsis, E. picta and Hemieleotris latifasciata (Gobiiformes: Eleotridae). Mitochondrial DNA B Resour. 2:747–750.
- Bacon CD, Silvestro D, Jaramillo C, Smith BT, Chakrabarty P, Antonelli A. 2015a. Biological evidence supports an early and complex emergence of the Isthmus of Panama. Proc Natl Acad Sci USA. 112:6110–6115.
- Bacon CD, Silvestro D, Jaramillo C, Smith BT, Chakrabarty P, Antonelli A. 2015b. Reply to Lessios and Marko et al.: Early and progressive migration across the Isthmus of Panama is robust to missing data and biases. Proc Natl Acad Sci USA. 112:E5767–E5768.
- Bagley JC, Alda F, Breitman MF, Bermingham E, van den Berghe EP, Johnson JB, Russello MA. 2015. Assessing species boundaries using multilocus species delimitation in a morphologically conserved group of Neotropical freshwater fishes, the Poecilia sphenops species complex (Poeciliidae). PLoS One. 10:e0121139.
- Coates AG, Obando JA. 1996. The geological evolution of the Central American Isthmus. In: Jackson J, Budd AF, Coates AG, editors. Evolution and environment in tropical America. Chicago (IL): University of Chicago Press; p. 21–56.
- Faircloth BC. 2013. illumiprocessor: a trimmomatic wrapper for parallel adapter and quality trimming. http://dx.doi.org/10.6079/J9ILL.
- Faircloth BC, McCormack JE, Crawford NG, Harvey MG, Brumfield RT, Glenn TC. 2012. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst Biol. 61:717–726.
- Faircloth BC, Sorenson L, Santini F, Alfaro ME. 2013. A phylogenomic perspective on the radiation of ray-finned fishes based upon targeted sequencing of Ultraconserved Elements (UCEs). PLoS One. 8:e65923.
- Galván-Quesada S, Doadrio I, Alda F, Perdices A, Reina RG, García Varela M, Hernández N, Campos Mendoza A, Bermingham E, Domínguez-Domínguez O, Bernardi G. 2016. Molecular phylogeny and biogeography of the amphidromous fish genus Dormitator Gill 1861 (Teleostei: Eleotridae). PLoS One. 11:e0153538.
- Guimarães-Costa A, Vallinoto M, Giarrizzo T, Angulo A, Ruiz-Campos G, Schneider H, Sampaio I. 2017. Exploring the molecular diversity of Eleotridae (Gobiiformes) using mitochondrial DNA. J Appl Ichthyol. 33:572–578.
- Jaramillo C, Montes C, Cardona A, Silvestro D, Antonelli A, Bacon CD. 2017. Comment (1) on ‘Formation of the Isthmus of Panama’ by O'Dea et al. Sci Adv. 3:e1602321.
- Jordan JS. 1908. The law of the geminate species. Am Nat. 42:73–80.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Knowlton N, Weigt LA. 1998. New dates and new rates for divergence across the Isthmus of Panama. Proc R Soc B Biol Sci. 265:2257–2263.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359.
- Lessios HA. 2008. The great American schism: divergence of marine organisms after the rise of the Central American Isthmus. Annu Rev Ecol Evol Syst. 39:63–91.
- Lessios HA. 2015. Appearance of an early closure of the Isthmus of Panama is the product of biased inclusion of data in the metaanalysis. Proc Natl Acad Sci USA. 112:E5765.
- Marko PB. 2002. Fossil calibration of molecular clocks and the divergence times of geminate species pairs separated by the Isthmus of Panama. Mol Biol Evol. 19:2005–2021.
- McMahan CD, Davis MP, Domínguez-Domínguez O, García-de-León FJ, Doadrio I, Piller KR, Rocha L. 2013. From the mountains to the sea: phylogeography and cryptic diversity within the mountain mullet, Agonostomus monticola (Teleostei: Mugilidae). J Biogeogr. 40:894–904.
- Miura O, Torchin ME, Bermingham E. 2010. Molecular phylogenetics reveals differential divergence of coastal snails separated by the Isthmus of Panama. Mol Phylogenet Evol. 56:40–48.
- Montes C, Cardona A, McFadden R, Moron SE, Silva CA, Restrepo-Moreno S, Ramirez DA, Hoyos N, Wilson J, Farris D, et al. 2012. Evidence for middle Eocene and younger land emergence in central Panama: Implications for Isthmus closure. Geol Soc Am Bull. 124:780–799.
- Myers GS. 1949. Salt-tolerance of fresh-water fish groups in relation to zoogeographical problems. Bijdr Tot De Dierk. 28:315–322.
- O’Dea A, Lessios HA, Coates AG, Eytan RI, Restrepo-Moreno SA, Cione AL, Collins LS, de Queiroz A, Farris DW, Norris RD. 2016. Formation of the Isthmus of Panama. Sci Adv. 2:e1600883.
- Pereira LHG, Hanner R, Foresti R, Oliveira C. 2013. Can DNA barcoding accurately discriminate megadiverse Neotropical freshwater fish fauna? BMC Genetics. 14:20.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Thacker CE. 2009. Phylogeny of Gobioidei and placement within Acanthomorpha, with a new classification and investigation of diversification and character evolution. Copeia. 2009:93–104.
- Thacker CE. 2017. Patterns of divergence in fish species separated by the Isthmus of Panama. BMC Evol Biol. 17:111.
- Thacker CE, Hardman MA. 2005. Molecular phylogeny of basal gobioid fishes: Rhyacichthyidae, Odontobutidae, Xenisthmidae, Eleotridae (Teleostei: Perciformes: Gobioidei). Mol Phylogenet Evol. 37:858–871.
- Ward RD. 2009. DNA barcode divergence among species and genera of birds and fishes. Mol Ecol Resour. 9:1077–1085.
