Abstract
In this study, the complete mitochondrial genome (mitogenome) of Oberthueria lunwan Zolotuhin & Wang, Citation2013 (Lepidoptera: Oberthuerinae) is reported for the first time. The entire mitogenome is a circular DNA molecule of 15,673 bp in length (GenBank accession number: MF100143), consisting of 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs) genes, two ribosomal RNAs (rRNAs) genes, and a control region (A + T-rich region). The phylogenetic trees are based on 13 PCGs amino acid sequences of 31 related lepidopteran species in which mitogenome sequences were constructed. The Oberthuerinae consisting of O. lunwan+Andraca theae was strongly supported as a monophyletic clade by the posterior probability of 1.00 and the bootstrap value of 100%, but the relationships among Oberthuerinae, Bombycidae, Satruniidae, and Sphingidae are need to be further confirmed.
The Oberthueria species are difficult for distinguishing from each other in general appearance (Zolotuhin and Wang Citation2013) with their complete mitochondrial genome (mitogenome) unreported. Oberthueria lunwan Zolotuhin and Wang, Citation2013 (Lepidoptera: Oberthuerinae) was previously recorded just from mainland China (Yunnan) and northeastern Myanmar with the larval host unknown (Wang et al. Citation2015). Here, the adults of this species were collected from Shennonggu National Forest Park (26°18′ 00″ N, 113°56′ 30″ E, 900 m at attitude), Yanling County, Hunan Province, Centre China on 12 September 2015. The genetic characters of O. lunwan are very helpful for understanding its phylogenetic relationships in the superfamily Bombycoidea and identifying the Oberthueria species correctly.
The mitogenome DNA was extracted from the adult’s thorax muscles and purified by using Wizard Genomic DNA Purification Kit (Promega, Beijing, China). All the adult specimens and the genomic DNA were deposited in Insect Museum of Hunan Agricultural University. Primers reported by Gu et al. (Citation2016) were used to amplify the complete mitogenome. The fragments were proof-read by using the software of Geneious version 8.1.2 (Kearse et al. Citation2012). The gene annotation was accomplished with MITOS (http://mitos.bioinf.uni-leipzaig.de) (Bernt et al. Citation2013). Filtrating the nonconservative parts was done using Gblocks 0.91b (Castresana Citation2000). The phylogenetic trees of the Bombycoidea with Lasiocampidae species as outgroups were established by using MrBayes 3.1.2 (Ronquist et al. Citation2012) with running for 5,000,000 generations and using raxmlGUI 1.5 program (https://sourceforge.net/projects/raxmlgui/) (Silvestro and Michalak Citation2012) with 1000 replications.
The complete mitogenome of O. lunwan is a circular DNA molecule of 15,673 bp in length, consisting of 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs) genes, two ribosomal RNAs (rRNAs) genes, and a control region (A + T-rich region). The mitogenome contains A (40.4%), T (39.3%), C (12.4%), and G (7.9%), which showed a high A + T bias. All of the PCGs started with ATN as the start codon except for cox1, which started with CGA. In addition, 11 of the 13 PCGs ended with TAA, whereas cox1 and cox2 ended with a single T. The A + T-rich region of the full-length 551 bp, located between rrnS and trnM gene, had 93.28% A + T content. The conserved region included an ‘ATAGT’ structure followed by poly-T. There were two microsatellites, ‘(TA)6’ and ‘(TA)8’, which were located at 86 bp and 202 bp upstream of rrnS, respectively.
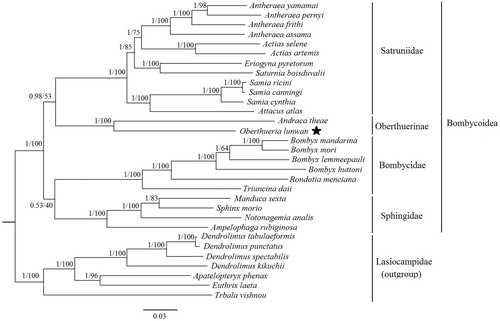
Based on the amino acid sequences of the 13 PCGs from 31 related lepidopteran species (Wang et al. Citation2015; Gu et al. Citation2016; Liu et al. Citation2017; Wu et al. Citation2017; Zhao et al. Citation2017), the phylogenetic trees were established with the 24 Bombycoidea species as ingroups and 7 Lasiocampidae species as outgroups. The maximum likelihood (ML) and Bayesian inference (BI) trees showed the same topological structures (). The Oberthuerinae consisting of the species Andraca theae and the O. lunwan formed a monophyletic clade supported by the bootstrap value of 100% and the posterior probability of 1.00. The phylogenetic relationships among Sphingidae, Bombycidae, Satruniidae, Oberthuerinae in Bombycoidea were uncertain with further studies necessary.
Figure 1. Bayesian inference and maximum likelihood phylogram constructed using 13 PCGs of mitogenomes with partitioned models. Numbers above each node indicate the ML bootstrap support values and the BI posterior probability. All the species’ accession numbers in this study are listed as below: Actias artemis KF_927042, Actias selene NC_018133, Ampelophaga rubiginosa NC_035431, Andraca theae KX365419, Antheraea assama NC_030270, Antheraea frithi NC_027071, Antheraea pernyi NC_004622, Antheraea yamamai NC_012739, Apatelopteryx phenax KJ508055, Attacus atlas NC_021770, Bombyx huttoni NC_026518, Bombyx lemeepauli KY620670, Bombyx mandarina NC_003395, Bombyx mori NC_002355, Dendrolimus kikuchii MF100138, Dendrolimus punctatus NC_027156, Dendrolimus spectabilis NC_025763, Dendrolimus tabulaeformis NC_027156, Eriogyna pyretorum NC_012727, Euthrix laeta NC_031507, Manduca sexta NC_010266, Oberthueria lunwan MF100143, Rondotia menciana NC_021962, Samia canningi NC_024270, Samia cynthia KC812618, Samia ricini NC_017869, Saturnia boisduvalii NC_010613, Sphinx morio NC_02078, Notonagemia analis KU934302, Trabala vishnou KU884483, Triuncina daii KY091643.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler P. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Gu XS, Ma L, Wang X, Huang GH. 2016. Analysis on the complete mitochondrial genome of Andraca theae (Lepidoptera: Bombycoidea). J Insect Sci. 16:105.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Liu SQ, Gu XS, Wang X. 2017. The complete mitochondrial genome of Bombyx lemeepauli (Lepidoptera: Bombycidae) and its phylogenetic relationship. Mitochondrial DNA B. 2:518–519.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RaxML. Org Divers Evol. 12:335–337.
- Wang X, Wang M, Zolotuhin VV, Hirowatari T, Wu S, Huang GH. 2015. The fauna of the family Bombycidae sensu lato (Insecta, Lepidoptera, Bombycoidea) from Mainland China, Taiwan and Hainan Islands. Zootaxa. 3989:1–138.
- Wu YH, Gu SX, Xue J, Wang X. 2017. The complete mitochondrial genome of Dendrolimus kikuchii (Lepidoptera: Lasiocampidae). Mitochondrial DNA B. 2:536–537.
- Zhao YP, Gu XS, Ren GF, Wang X. 2017. The complete mitochondrial genome of Triuncina daii (Lepidoptera: Bombycidae) and its phylogenetic implications. Entomotaxonomia. 39:223–237.
- Zolotuhin VV, Wang X. 2013. A taxonomic review of Oberthueria Kirby, 1892 (Lepidoptera, Bombycidae: Oberthuerinae) with description of three new species. Zootaxa. 3693:465–478.
