Abstract
Diaphanosoma has been called “tropical Daphnia” for its strong ecological role in tropical freshwater as Daphnia in temperate waters. The present study sequenced and annotated the mitochondrial genome (MG) of Diaphanosoma dubium. The MG of Diaphanosoma dubium is 16,362 bp in length, with typical metazoan gene composition. Phylogenetic analysis confirms an earlier finding that Neodiaphanosoma can be separated from Diaphanosoma as a subgenera. One unknown extra CDS region and different arrangement of tRNA were identified when this MG was compared to that of Daphnia magna. This is the first non-daphnia MG of Cladocera, and information on MG sequence and tRNA order provide valuable molecular data in understanding phylogeny of Diaphanosoma and Cladocera.
Daphnia is the keystone genus in temperate aquatic ecosystems, and its tropical partner, Diaphanosoma, plays a similar role in warm waters and has been called “tropical Daphnia” (Dumont Citation1994; Sarma et al. Citation2005). Among all Diaphanosoma, Diaphanosoma dubium is one of the most widely distributed species (Korovchinsky Citation2000; Liu et al. Citation2018), and it is the common dominant Cladocera in eutrophic lakes and reservoirs in tropics and subtropics (Chen et al. Citation2011). Its high population density could indicate a strong ecological function. Therefore, Diaphanosoma dubium can be a model organism to study effect of global change in tropics and population genetics over large spatial scale. However, few molecular data are available for Diaphanosoma dubium and its congeners. Mitochondrial genomes (MG) can increase resolution in phylogeny (Cameron Citation2014; Schreeg et al. Citation2016) and provide more genetic information when compared to the frequently used barcoding sequence (COI). Here, we sequenced and annotated the MG of Diaphanosoma dubium, and compared it to that of Daphnia magna.
Living animal was collected from Liuxihe reservoir (116.20 E, 39.97 N) and massively cultured in small aquarium. Sample is preserved in Institute of Hydrobiology, Jinan University, Guangzhou, China. Thousands of individual was gathered after 1 moth of cultivation, and the collection was preserved in −80 °C before sending to Beijing Genomics Institute (BGI) for next-generation sequencing (Hiseq 2500, PE-150). The assembly and annotation procedure followed Xu et al. (Citation2017) with COI sequence of Diaphanosoma dubium (AB549201) as seed. COI of four other Diaphanosoma species (D. excisum, D. orghidani, D. brachyurum and D. mongolianum) were amplified with 2 pairs of primers: LCO1490 and HCO2198 (Vrijenhoek Citation1994), CrustF (Costa et al. Citation2007) and HCO2198. Based on COI sequence of all available Sididae species from NCBI and this study, a maximum-likelihood phylogeny was reconstructed with PhyML (Guindon et al. Citation2010) in Seaview (Gouy et al. Citation2010). Finally, the synteny of all mitochondrial genes between Diaphanosoma dubium and Daphnia magna was analyzed using SimpleSynteny (Veltri et al. Citation2016).
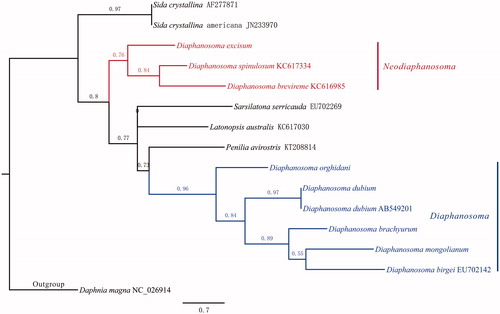
The total length of Diaphanosoma dubium MG (MG428405) is 16,362 bp, with A + T biased composition: A (27.8%), C (16.1%), G (18.2%), T (37.8%). Thirty-seven genes were annotated: 13 protein coding genes, 22 tRNA genes and two rRNA genes. Among all PCGs, ND2 and ND6 are initiated with ATT, COX1 with GTG, and the rest with ATG. Twelve genes end up with complete stop codon: ND2, COX1 and CYTB with TAG, ATP6, ATP8, COX3, ND1, ND3, ND5, ND4, ND4L and ND6 with TAA. Only COX2 end up with a single “T”. tRNA genes have length ranging from 58 to 72 bp, and they are folded into typical cloverleaf structure. The putative control region is 517 bp in length, with high A + T composition (63.4%). The phylogeny for Sididae was in compatible with that from Guo (Citation2015) (). Diaphanosoma was split into two subgenera, namely Diaphanosoma Fischer S.S. and Neodiaphanosoma Paggi. When compared with the MG of Daphnia magna (NC_026914), Diaphanosoma dubium’s MG is 1414 bp longer, which could mostly result from an extra unknown CDS (locate between 1496 and 2473 bp). Moreover, different arrangement of tRNA was found between these two species, and no shift of PCGs was discovered.
Figure 1. Phylogeny of Sididae. Sequence without NCBI ID are from this study. Parameters in PhyML were set as following: aLRT for Branch Support, optimize nucleotide equilibrium frequencies, invariable site ratio, and selected best of NNI & SPR for tree searching operation, all the rest are left as default.
Disclosure statement
None of the authors has any conflict of interest to declare. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 2014. 59:95–117.
- Chen H, Cheng D, Xu L, Lin Q, Han B. 2011. Distribution of Diaphanosoma dubium and D. orghidani in reservoirs of Guangdong Province, southern China. J Lake Sci. 23:801–805.
- Costa FO, deWaard JR, Boutillier J, Ratnasingham S, Dooh RT, Hajibabaei M, Hebert PDN. 2007. Biological identifications through DNA barcodes: the case of the Crustacea. Can J Fish Aquat Sci. 64:272–295.
- Dumont HJ. 1994. On the diversity of the Cladocera in the tropics. Hydrobiologia. 272:27–38.
- Gouy M, Guindon S, Gascuel O. 2010. SeaView Version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27:221–224.
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321.
- Guo F. 2015. Towards a phylogenetic taxonomy of the genus Diaphanosoma (Crustacea, Ctenopoda, Sididae) based on the sixth trunk limb and on COI gene sequences. Guangzhou: Jinan University.
- Korovchinsky NM. 2000. Redescription of Diaphanosoma dubium Manuilova, 1964 (Branchiopoda: Ctenopoda: Sididae), and description of a new, related species. Hydrobiologia. 441:73–92.
- Liu P, Xu L, Xu S, Martínez A, Chen H, Cheng D, Dumont HJ, Han B-P, Fontaneto D. 2018. Species and hybrids in the genus Diaphanosoma Fisher, 1850 (Crustacea: Branchiopoda: Cladocera). Mol Phylogenet Evol. 118:369–378.
- Sarma SSS, Nandini S, Gulati RD. 2005. Life history strategies of cladocerans: comparisons of tropical and temperate taxa. Hydrobiologia. 542:315–333.
- Schreeg ME, Marr HS, Tarigo JL, Cohn LA, Bird DM, Scholl EH, Levy MG, Wiegmann BM, Birkenheuer AJ. 2016. Mitochondrial genome sequences and structures aid in the resolution of piroplasmida phylogeny. Plos One. 11:e0165702
- Veltri D, Wight MM, Crouch JA. 2016. SimpleSynteny: a web-based tool for visualization of microsynteny across multiple species. Nucleic Acids Res. 44:W41–W45.
- Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Marine Biol Biotechnol. 3:294–299.
- Xu S, Guan Z, Huang Q, Xu L, Vierstraete A, Dumon H, Lin Q. 2017. The mitochondrial genome of Atrocalopteryx melli Ris, 1912 (Zygoptera: Calopterygidae) via Ion Torrent PGM NGS sequencing. Mitochondrial DNA B. DOI: 10.1080/23802359.2017.1413307.
