Abstract
The complete mitochondrial genome of Gerres filamentosus was sequenced by high throughput sequencing method. Length of this genome is 16,795 bp, containing 13 protein-coding genes, 22 tRNA genes, two rRNA genes and one large non-coding region. ND6 and eight tRNA genes are encoded by L-strand, and others are encoded by H-strand, which is similar to those in most vertebrates. Phylogenetic tree based on 13 protein-coding genes shows that the clade of G. filamentosus is closely clustered with that of Gerres oyena, and families Caproidae and Ephippidae have the closest relationship to Gerridae, comparing with Sillaginidae
Gerridae fishes are marine species (occasionally brackish and rarely in freshwater) inhabiting in warm seas. Gerres filamentosus is widely distributed in tropical and sub-tropical waters of worldwide. Gerridae species has been confused in taxonomic status for long time, because of their similar morphological characters and coloration (Heemstra and Yukio Citation2007). In this paper, we got the sample from Naozhou island in Zhanjiang (geographic coordinate: N 20°53′20.11″, E 112°28′46.2″). The whole body specimen was preserved in ethanol and registered to the Marine Biodiversity Collection of South China Sea, Chinese Academy of Sciences, under the voucher number SCF20171022002. It is the first reported about the complete mitogenome of G. filamentosus, and analyzed its phylogenetic relationships within order Perciformes.
The complete mitochondrial genome of G. filamentosus has 16,795 bp in length (GenBank accession no. MG386479), including 13 protein-coding genes, two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes and one control region (D-Loop). Genes encoding on the genome are similar to other vertebrates (Yu and Kwak Citation2015). The OL (origin of replication on the light-strand) was found in the cluster of five tRNA genes (WANCY region) between tRNA-Asn and tRNA-Cys. And the D-Loop (control region) is 956 bp in length, between tRNA-Pro and tRNA-Phe. Among 13 protein-coding genes, 12 ORF started with ATG and rest one (COI) started with GTG, which is as same as Liobagrus styani (Huang et al. Citation2017). However, for the stop codon, ND4 and Cytb ended with just a single base T, which is different from the common stop codon with three bases. But the same situation was also found in many other fishes, such as Liobagrus styani (Huang et al. Citation2017), Brachymystax lenok tsinlingensis (Yu and Kwak Citation2015). Among above genes, except ND6 and eight tRNA genes were encoded on L-strand, others were encoded on H-strand, which is similar to those of other vertebrates. The base composition of genome of G. filamentosus is T: 25.9%, C: 30.0%, A: 27.6% and G: 16.5%.
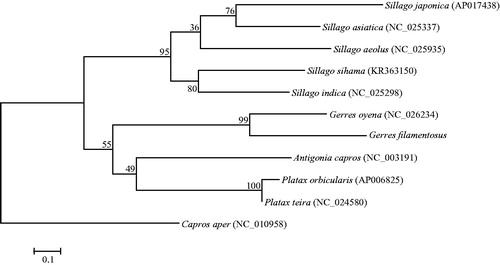
The phylogenetic relationships of G. filamentosus with 10 closely related species were analyzed in this study. The complete mitochondrial genes of these ten species are available on GenBank. The maximum-likelihood evolutionary tree (ML tree) was constructed by MEGA 7 (Kumar et al. Citation2016) based on 1st and 2nd codon sequences of 13 protein-coding genes. In the ML tree, G. filamentosus clustered with G. oyena with a strong support, A. capro, P. orbicularis and P. teira formed a clade as a sister lineage to clade of G. filamentosus and G. oyena (). These results show that Caproidae and Ephippidae have closer phylogenetic relationship to Gerridae than to Sillaginidae.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Heemstra PC, Yukio I. 2007. A new Gerreid fish species and redescription of Gerres maldivensis Regan, 1902 from the Indian Ocean (Perciformes: Gerreidae). Copeia. 2007:85–92.
- Huang JY, Hu S, Bai X, Zhang E. 2017. Complete mitochondrial genome of Liobagrus styani (Teleostei: Amblycipitidae). Mitochondrial DNA Part B Resources. 2:15–16.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Yu JN, Kwak M. 2015. The complete mitochondrial genome of Brachymystax lenok tsinlingensis (Salmoninae, Salmonidae) and its intraspecific variation. Gene. 573:246–253.