Abstract
This is a de novo assembly and annotation of a complete mitochondrial genome from Pyrus pyrifolia in the family Rosaceae. The complete mitochondrial genome of P. pyrifolia was assembled from PacBio RSII P6-C4 sequencing reads. The circular genome was 458,873 bp in length, containing 39 protein-coding genes, 23 tRNA genes and three rRNA genes. The nucleotide composition was A (27.5%), T (27.3%), G (22.6%) and C (22.6%) with GC content of 45.2%. Most of protein-coding genes use the canonical start codon ATG, whereas nad1, cox1, matR and rps4 use ACG, mttB uses ATT, rpl16 and rps19 uses GTG. The stop codon is also common in all mitochondrial genes. The phylogenetic analysis showed that P. pyrifolia was clustered with the Malus of Rosaceae family. Maximum-likelihood analysis suggests a clear relationship of Rosids and Asterids, which support the traditional classification.
Pears (Pyrus spp.), which belong to the family of Rosaceae, have been cultivated in East Asia, and in other countries such as India, Australia, New Zealand, and the United States for more than 3000 years, and are one of most important fruit crops in temperate regions (Bell Citation1991). Pyrus accessions can be classified into three groups. One group comprises P. brestchenidri, P. pyrifolia, and P. ussureinsis in the commercial cultivation of Asia. And also, there is European pear (P. communis), the other rootstock group contains P. betulaefolia and P. calleryana. Asian pears are classified in first of these groups and have a sweet flavour, rich juice, and crisp flesh (Kim Citation2016).
The plant sample of Wonwhang (Kim et al. Citation1995) was collected from Naju (Latitude: 35° 01′25.6″N, Longitude: 126° 44′38.4″E), Republic of Korea and representatively identified by Dr Y. K. Kim of the Pear Research Station, National Institute of Horticultural & Herbal Science, Rural Development Administration. Total genomic DNA was extracted from 1 g of young leaves using 1 ml aqua phenol (Mpbio 11AQUAPH) following Kim et al. (Citation2006) with minor modifications. The complete chloroplast genome of Wonwhang, Pyrus pyrifoia, was assembled and analyzed by de novo assembly using whole-genome sequencing data (Chung et al. Citation2017). Sequencing was performed using a PacBio RSII platform in P6-C4 chemistry (Pacific Biosciences, Menlo Park, CA) at DNA Link (http://www.dnalink.com/korean/) in Seoul, Republic of Korea. The Single Molecule, Real-Time (SMRT) sequencing, PacBio RSII P6-C4 sequencing, was conducted using the smrtmake assembly pipeline (Chin et al. Citation2013). We performed the de novo assembly using Canu (Koren et al. Citation2016) to produce a complete mitochondrial genome. And ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) were predicted using RNAmmer 1.2 (Lagesen et al. Citation2007) and tRNAscan-SE 1.4 (Lowe and Eddy Citation1997). The annotated mitochondrial genome sequence using MITOS (Bernt et al. Citation2013) was deposited into GenBank under the accession number KY563267.
The circular genome of P. pyrifolia is 458,873 bp in length, which consists of 39 protein-coding genes (PCGs), 23 tRNAs and three rRNAs. The nucleotide contents of A, T, G and C in mitochondrial genome were 27.5%, 27.3%, 22.6% and 22.6%, respectively. Most of PCGs contained ATG as the start codon, while nad1, cox1, matR and rps4 began with ACG, rpl16 and rpl19 with GTG and mttB started with ATT. All PCGs were terminated with the typical stop codons. The 23 tRNA genes ranged from 71 bp and 88 bp in length. The rrn5, rrnS and rrnL were 119 bp, 1782 bp and 3131 bp, respectively.
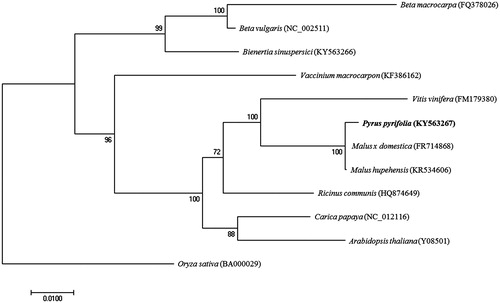
Phylogenetic analysis was performed using the complete mitochondrial genomes of twelve members including seven Rosids, one Asterids and three Amaranthaceae family, Oryza sativa in Monocut was used as outgroup. Sequences for each mitochondrial genome were aligned using ClustalW, and a maximum-likelihood phylogenetic tree was reconstructed using MEGA7 (Kumar et al. Citation2016) with 1000 bootstrap replicates (). The phylogenetic tree showed that mitochondrial genome of P. pyrifolia was clustered with Malus genus and belonged to Rosids clade. The complete mitochondrial genome of P. pyrifolia will provide essential and important DNA molecular data for phylogenetic and evolutionary analysis for Asian pears.
Figure 1. Maximum-likelihood tree of complete mitochondrial genome of P. pyrifolia and 11 other species. The numbers at the nodes are bootstrap percent probability values based on 1000 replications. Mitochondrial genome sequences were obtained from GenBank and their accession numbers are indicated next to species name.
Disclosure statement
The authors declare no conflicts of interest.
Additional information
Funding
References
- Bell RL. 1991. Pears (Pyrus). Genetic Resources of Temperate Fruit and Nut Crops. 290:657–700.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Chin C, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nature Methods. 10:563–569.
- Chung HY, Lee TH, Kim YK, Kim JS. 2017. Complete chloroplast genome sequences of Wonwhang (Pyrus pyrifolia) and its phylogenetic analysis. Mitochondrial DNA Part B. 2:325–326.
- Kim HC, Hwang HS, Shin YE, Lee DG, Kang SJ, Shin IS, Cheon BD, Moon JY, Kim JH, Kim SB. 1995. Breeding of a new early large sized high-quality pear cultivar “Wonwhang”. Korean J Agricult Sci. 37:471–477.
- Kim JS, Chung TY, King GJ, Jin M, Yang TJ, Jin YM, Kim HI, Park BS. 2006. A sequence-tagged linkage map of Brassica rapa. Genetics. 174:29–39.
- Kim YK. 2016. Diversity of Pyrus germplasm and breeding of red skin colored and scab resistant pear by using interspecific hybridization. PhD degree, Chonnam National University.
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27:722-736.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 7:3100–3108.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
