Abstract
Acorssocheilus beijiangensis is an endemic south China stream-dwelling cyprinid species. Its complete mitochondrial genome is 16,596 bp in length, consisting of 13 protein-coding genes, 22 tRNA genes (ranging from 67 bp in tRNACys to 76 bp in tRNALeu and tRNALys), two rRNA genes (959 bp in 12S rRNA and 1683 bp in 16S rRNA), and one control region (937 bp). Its overall base composition is A: 31.1%, C: 27.9%, G: 16.2%, and T: 124.8%. The complete mitogenome of the Chinese barred species of Cyprinidae could provide a basic data for further phylogenetics analysis.
Acrossocheilus beijiangensis is a barred cyprinid species from the Pearl River (Zhu-Jiang) of Guizhou, Guangxi and Guangdong provinces in China (Wu et al. Citation1977; Shan et al. Citation2000). This species differs from the congenus with a combination of characters: five vertical bars on the flanks 2–4 scales wide extending slightly beyond the lateral line; the second bar positioned posterior to the base of the last simple dorsal-fin ray; no longitudinal stripe along the lateral line; two thick lateral lobes of the lower lip; last unbranched dorsal-fin ray stout with a serrated posterior edge; membranes between the dorsal-fin rays oblong with black blotches. However, ontogenetic changes and/or sexual dimorphism in body coloration and mouthpart structures usually confuse researchers when delineating these barred species. Thus, combination of mitochondrial DNA data and morphological traits will help for the species identification of taxonomic validity and phylogenetic position of the species.
The sample caught from the Congjiang county (Duliu-Jiang, flowing to the Xi-Jiang River) of Guizhou Province in China was deposited in the collection of the Zhejiang Museum of Natural History under the accession number ZMNH 2011081501. The complete mitogenome of Acorssocheilus beijiangensis has been obtained from high-throughput sequencing on whole-genomic DNA with HiSeq 2000 platform (Illumina, San Diego, CA). We used next-generation sequencing to perform low-coverage whole-genome sequencing according to previous protocol (Shen et al. Citation2016). The complete mitogenome of A. beijiangensis is 16,596 bp in length (GenBank KY131976), includes 13 protein-coding genes, 22 tRNA genes (ranging from 67 bp in tRNACys to 76 bp in tRNALeu and tRNALys), two rRNA genes (959 bp in 12S rRNA and 1683 bp in 16S rRNA) and one D-loop control region (937 bp). Its overall base composition is A: 31.1%, C: 27.9%, G: 16.2%, and T: 24.8% which shows AT bias, with the AT content of 55.9%. The complete mitogenome of A. beijiangensis showing 97% identities to A. stenotaeniatus (GenBank KJ909660) after BLAST search against NCBI nr/nt database.
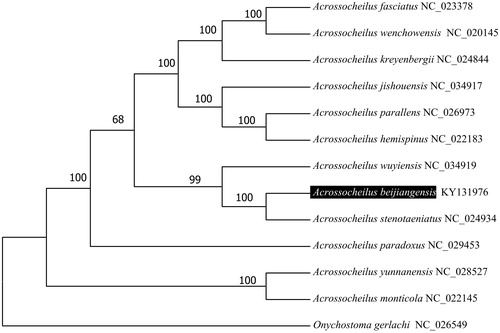
To validate the phylogenetic position of A. beijiangensis, we used MEGA6 software (Tamura et al. Citation2013) to construct a maximum-likelihood tree (with 100 bootstrap replicates and Kimura 2-parameter model) containing complete mitogenomes of 12 species derived from Acrossocheilus genus. Onychostoma gerlachi (Cheng et al. Citation2016) derived from Onychostoma genus was used as outgroup for tree rooting (). The resultant phylogeny shows that the A. beijiangensis is closely related to A. stenotaeniatus with high bootstrap value supported. The Kimura-two-parameter distance was 0.029 between A. beijiangensis and A. stenotaeniatus and was 0.074 between A. beijiangensis and A. wuyiensis. The complete mitogenome of A. beijiangensis which deduced in this study provides a basic data for further phylogenetic and conversational analysis of the Chinese barred species in Cyprinidae.
Figure 1. Molecular phylogeny of Acrossocheilus beijiangensis and related species based on complete mitogenome. The complete mitogenomes is downloaded from GenBank and the phylogenic tree is constructed by maximum-likelihood method with 500 bootstrap replicates. The gene's accession number for tree construction is listed behind the species name.
Acknowledgements
Our sincere thanks to Zhuo-Cheng Zhou and Xiao-Lu Yu for helping with field work. We also thank Dr. Chung-Der Hsiao from Chung Yuan Christian University in Taiwan for his help on Bioinformatic analysis of NGS data.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Cheng Q, Chen F, Huang DM, Wang HY. 2016. The complete mitochondrial genome of the Onychostoma gerlachi (Cypriniformes: Cyprinidae). Mitochondrial DNA A. 27:3210–3211.
- Shan X, Lin R, Yue P, Chu X. 2000. Barbinae. In: Yue PQ, editors. Fauna Sinica (Osteichthyes: Cypriniformes III). Beijing: Science Press; pp. 126–147. [In Chinese]
- Shen KN, Yen TC, Chen CH, Li HY, Chen PL, Hsiao CD. 2016. Next generation sequencing yields the complete mitochondrial genome of the flathead mullet, Mugil cephalus cryptic species NWP2 (Teleostei: Mugilidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1758–1759.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wu H, Lin R, Chen Q. 1977. Barbinae. In: Wu HW, editor. The cyprinid fishes of China. Part 2. Shanghai: Shanghai People’s Press; pp. 229–394. [In Chinese]
