Abstract
The Chinese pepper (Zanthoxylum simulans) is a flowering plant in the family Rutaceae, native to eastern China and Taiwan. Like many other members of the Rutaceae, it is an economically important aromatic crop known for its volatile oil and nutrition. We determined the complete chloroplast genome sequences for Z. simulans using IIumina sequencing. The Z. simulans chloroplast has a total length of 158,461 bp; it consists of a large single copy (LSC) region of 85,568 bp, a small single copy region (SSC) length of 17,603 bp, and an inverted region (IR) of 27,645 bp. The genome encodes 132 annotated genes, including 87 protein-coding genes, 37 tRNA, and 8 rRNA. The overall GC content of the Z. simulans chloroplast genome was 38.5%. A phylogenomic analysis revealed that Z. simulans are clustered with Z. bungeanum within the genus Zanthoxylum.
The Zanthoxylum simulans is an angiospermae plant in the family Rutaceae, endemic to eastern China and Taiwan (Feng et al. Citation2016). It is one of several species of Zanthoxylum from which Sichuan pepper is produced, which is a commonly used spice in Desi Chinese cuisine, Chinese, Korea, Tibetan, Nepali, and Indian cuisine (Paik et al. Citation2005). The husk or hull (pericarp) around the seeds may be used whole, especially in Sichuan cuisine or Szechwan cuisine or Szechuan cuisine, and the finely ground powder is one of the ingredients for five-spice powder (Lee et al. Citation2016). The genus Zanthoxylum includes more than 45 species and 13 varieties in China (Feng et al. Citation2016).
Here, we assembled the complete chloroplast genome sequence of Z. simulans using the high-throughput Illumina paired-end sequencing data. The annotated plastid genome has been deposited into GenBank with the accession number: MF716524. Total genomic DNA was extracted from a single individual Z. simulans growing in Jiuzhou town Xianren Ba village, Guizhou, China (106°08′E, 26°16′N). DNA sample and voucher specimen of Z. simulans were deposited in the Botany Laboratory, Guizhou Academy of Forestry (Guiyang, Guizhou, China). After trimming the sequences, high-quality PE reads were aligned to the Z. bungeanum (NCBI Accession number: KX497031) (Liu and Wei Citation2017) using bowtie (Langmead and Salzberg Citation2012), and assembled into a genome using SPAdes assembler (Bankevich et al. Citation2012). Annotation was performed with Dual Organellar Genome Annotator (DOGMA) software (Wyman et al. Citation2004). We corrected the annotation with Geneious (Kearse et al. Citation2012).
The Z. simulans whole chloroplast genome is 158,461 bp in length. GC content was 38.5%. It contained large single copy (LSC) of 85,568 bp, a small single copy region (SSC) of 17,603 bp, and a pair of inverted repeat (IRs) of 27,645 bp. The genome encodes 132 functional genes, including 87 protein-coding genes, 37 tRNA, and 8 rRNA. The newly sequenced plastid genome is quite similar to other Zanthoxylum species in terms of overall organization, gene/intron content, gene order, and GC content (Lee et al. Citation2016; Liu and Wei Citation2017).
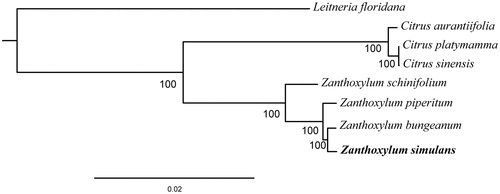
We performed a phylogenetic analysis based on the complete chloroplast genome of Z. simulans and six other species from the family Rutaceae, including three from the genus Zanthoxylum and three from the genus Citrus. Leitneria floridana (Simaroubaceae) was used as an outgroup. The Maximum Likelihood (ML) phylogenetic tree analysis was performed using RAxML (Stamatakis Citation2014). The phylogenetic tree showed that Z. simulans was most closely related to Z. bungeanum (). The newly characterized Z. simulans chloroplast genome can be used for studies of the evolutionary history of Zanthoxylum and to investigate genetic subdivision within the remaining Z. simulans germplasm resources.
Figure 1. Maximum Likelihood (ML) phylogenetic tree based on complete chloroplast genome sequences of six species from the family Rutaceae using L. floridana of Simaroubaceae as an outgroup. Numbers on branches are bootstrap support values based on 10,000 iterations. All eight species’s accession numbers are listed as below: Z. piperitum KT153018, Z. piperitum NC_027939, Z. schinifolium KT321318, C. aurantiifolia KJ865401, C. platymamma KR259987, C. sinensis NC_008334, and L. floridana NC_030482.
Disclosure statement
The authors declare no conflicts of interest.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Feng S, Liu Z, Chen L, Hou N, Yang T, Wei A. 2016. Phylogenetic relationships among cultivated Zanthoxylum species in China based on cpDNA markers. Tree Genet Genomes. 12:45–53.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359.
- Lee J, Lee HJ, Kim K, Lee SC, Sung SH, Yang TJ. 2016. The complete chloroplast genome sequence of Zanthoxylum piperitum. Mitochondrial DNA. A DNA Mapp Seq Anal. 27:3525–3526.
- Liu Y, Wei A. 2017. The complete chloroplast genome sequence of an economically important plant, Zanthoxylum bungeanum (Rutaceae). Conserv Genet Resour. 9:25–27.
- Paik SK, Koh KH, Beak SM, Paek SH, Kim JA. 2005. The essential oils from Zanthoxylum schinifolium pericarp induce apoptosis of HepG2 human hepatoma cells through increased production of reactive oxygen species. Biol Pharm Bull. 28:802–807.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
