Abstract
The flower shape of Lilium bulbiferum L. is different from that of the other European lilies. The phylogenetic position of the plant has been changed frequently. In the present study, we have sequenced the complete plastome of L. bulbiferum. The plastome sequence was 152,686 bp long, with a large single copy (LSC) region of 82,083 bp, a small single copy (SSC) region of 17,619 bp, and two inverted repeat (IR) regions of 26,492 bp each. It contained 133 genes, including 83 coding genes, 8 ribosomal RNAs, 38 transfer RNAs, and 4 pseudogenes. The GC contents of LSC, SSC, and IT were 34.8%, 30.7%, and 42.5%, respectively, corresponding to 37.0% of the total GC content. The phylogenetic position of L. bulbiferum was closely related to the north-eastern lilies.
Keywords:
Lilium bulbiferum L. is native to Europe, and is a rare plant in the wild (McRae et al. Citation1998; İkinci et al. Citation2006). This species is thus named because of the numerous bulbils in its leaf axils (McRae et al. Citation1998). Because of its erect bowl-shaped flower, Baker (Citation1871) and Wilson (Citation1925) distinguished L. bulbiferum from the other European lilies. However, Comber (Citation1949) reclassified the genus Lilium, and placed L. bulbiferum under the section Liriotypus, which included all European lilies, except L. martagon. Although many phylogenetic studies have been conducted on the genus Lilium, the phylogenetic position of L. bulbiferum remains unclear owing to its low phylogenetic resolution (Nishikawa et al. Citation2001; İkinci et al. Citation2006).
The complete plastome of L. bulbiferum was sequenced in the present study. Genomic DNA was extracted from the fresh leaves of L. bulbiferum by CTAB method (Doyle Citation1987), and it is stored in Kyungpook National University. Total DNA was sequenced using a HiSeq 2500 instrument (Illumina, San Diego, CA). The assembly strategy for mapping the plastome sequence followed the method of Kim et al. (Citation2015). The average depth of the plastome sequence was 458.2, with maximal and minimal depths of 759 and 152, respectively.
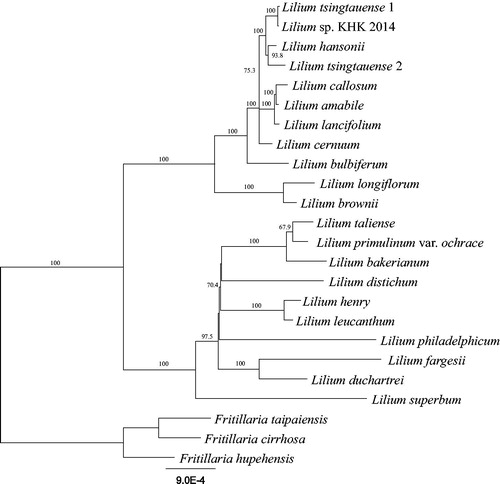
Gene annotation of plastome was performed using Geneious (Kearse et al. Citation2012) by comparing with the plastome sequences of other Lilium species. For phylogenetic analysis, plastome sequences of 20 Lilium and 3 Fritillaria species were downloaded from the NCBI data base, and a total of 77 genes were extracted from their plastome sequences. Each gene was aligned by MAFFT (Katoh et al. Citation2002), and the alignments were concatenated. The phylogenetic tree was constructed using RAxML (Stamatakis Citation2014) with GTR + G + I model in the CIPRES Science Gateway (Miller et al. Citation2010).
The plastome sequence of L. bulbiferum (GenBank accession number MG574829) was 152,686 bp long, with a large single copy (LSC) region of 82,083 bp, a small single copy (SSC) region of 17,619 bp, and two inverted repeat (IR) regions of 26,492 bp each. It contained 133 genes, including 83 coding genes, 8 ribosomal RNAs, 38 transfer RNAs, and 4 pseudogenes. Interestingly, the psaJ gene was extended by six bp, since its stop codon sequence TGA was substituted with TTA. The GC contents of LSC, SSC, and IR were 34.8%, 30.7%, and 42.5%, respectively, corresponding to 37.0% of the total GC content.
The phylogenetic analysis using 21 Lilium and 3 Fritillaria species showed that the genus Lilium can be distinguished into two main groups and L. bulbiferum was a sister clade of the north-eastern lilies (). The plastome sequence of L. bulbiferum is the first to be reported among the European lilies. Further, we expect that this sequence will help clarify the origin of European lilies, as well as new circumscription of the genus Lilium.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Baker J. 1871. A new synopsis of all the known lilies. Gard Chron. 28:104.
- Comber HF. 1949. A new classification of the genus Lilium. Lily Year book, Royal Horticultural Society. 13:85–105.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- İkinci N, Oberprieler C, Güner A. 2006. On the origin of European lilies: phylogenetic analysis of Lilium section Liriotypus (Liliaceae) using sequences of the nuclear ribosomal transcribed spacers. Willdenowia. 36:647–656.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kim HT, Kim JS, Moore MJ, Neubig KM, Williams NH, Whitten WM, Kim JH. 2015. Seven new complete plastome sequences reveal rampant independent loss of the NDH gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS One. 10:e0142215.
- McRae EA, Austin-Mcrae E, MacRae E. 1998. Lilies: a guide for growers and collectors. Portland: Timber Press.
- Miller M, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), IEEE.
- Nishikawa T, Okazaki K, Arakawa K, Nagamine T. 2001. Phylogenetic analysis of section Sinomartagon in genus Lilium using sequences of the internal transcribed spacer region in nuclear ribosomal DNA. Breeding Science. 51:39–46.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wilson E. 1925. The lilies of Eastern Asia, a monograph. London.