Abstract
The mitochondrial genome (mitogenome) of Polygonia c-aureum (Lepidoptera: Nymphalidae: Nymphalinae) is determined to be 15,209 bp in length and shows AT bias (80.6%). Similar to other butterflies, it contains 37 typical mitochondrial genes and one AT-rich region (D-loop). All protein-coding genes (PCGs) started with ATN, except for cox1 gene with CGA(R), which is often found in other butterflies, and seven PCGs harbour the typical stop codon TAA, whereas cox1, cox2, nad3, nad5, nad4 and nad1 end with a single T. The rrnL and rrnS genes are 1332 bp and 773 bp in length, respectively. The 342 bp AT-rich region contains non-repetitive sequences, but harbour several features common to the lepidopterans, including the motif ATAGA followed by a 19-bp poly-T stretch and a microsatellite-like (TA)8 element preceded by the ATTTA motif. The complete mitogenome sequence provided here would be useful for further understanding the taxonomy and phylogeny of Nymphalinae.
The subfamily Nymphalinae (Lepidoptera: Nymphalidae) comprises about 500 species and is distributed nearly all around the world (Harvey Citation1991). However, the taxonomy and phylogeny of Nymphalinae are still standing as a controversial issue, and need to be elucidated (Wahlberg et al. Citation2009; Shi et al. Citation2015). In recent decades, the insect mitochondrial genomes (mitogenomes) have been widely used as an informative molecular marker for phylogenetic and population genetic studies at various hierarchical levels, due to their unique features (Boore Citation1999; Salvato et al. Citation2008; Wu et al. Citation2014; Timmermans et al. Citation2016).
Here, we sequenced and characterized the complete mitogenome of Polygonia c-aureum (tribe Nymphalini). Adult individuals of P. c-aureum were netted at Nanjing, Jiangsu province, China (coordinates: E118°46’, N32°03’) on July 2012. After morphological identification, fresh individuals were preserved in 100% ethanol and kept in the laboratory at −20 °C under the accession number SQH-20120718. Total genomic DNA was extracted from the thorax muscle of a single individual using the Sangon Animal genome DNA Extraction Kit (Shanghai, China). The resultant reads were assembled and annotated using the BioEdit 7.0 (Hall Citation1999) and MEGA6 software (Tamura et al. Citation2013) with reference to mitogenome of Hypolimnas bolina (GenBank accession NC_026072) (Shi et al. Citation2015).
The complete mitogenome of P. c-aureum is a circular molecule of 15,209 bp in size (GenBank accession MF407452), containing typical insect 13 protein-coding genes (PCGs), two ribosomal RNA genes (rRNAs) and 22 transfer RNA genes (tRNAs), along with an AT-rich control region. Its gene arrangement and orientation are identical to those of other known nymphalid mitogenomes (e.g. Wu et al. Citation2014; Shi et al. Citation2015; McCullagh and Marcus Citation2015; Timmermans et al. Citation2016). The A + T content of the P. c-aureum mitogenome is 80.6%, which is generally in accordance with other nymphalid mitogenomes. All PCGs are initiated by typical ATN codons, except for cox1 gene, which is started with the unusual CGA(R) as observed in most of the other sequenced nymphalid butterflies (Kim et al. Citation2010; Hao et al. Citation2013; Gan et al. Citation2014; Shi et al. Citation2015). Seven PCGs have a complete stop codon (TAA), while cox1, cox2, nad3, nad5, nad4 and nad1 end with a single T. All tRNAs harbour the typical predicted secondary cloverleaf structures except for the trnS1, as seen in all other determined butterflies (Hao et al. Citation2013; Shi et al. Citation2015). The length of rrnL and rrns genes is 1332 bp and 773 bp, respectively. The putative AT-rich region is 342 bp long (93.8% A + T content) with several structures characteristic of lepidopterans.
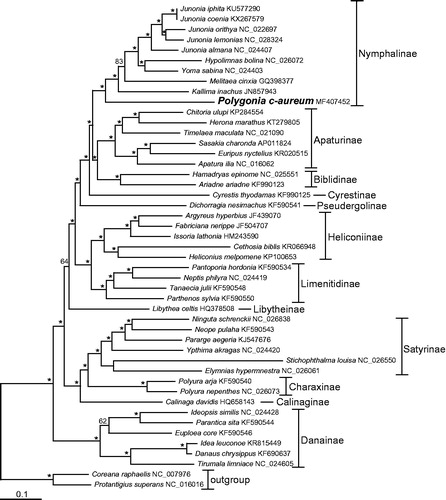
We performed a maximum-likelihood (ML) phylogenetic analysis with RAxML (version 7.2.6) using nucleotides sequences of 13 PCGs and two rRNAs to understand the phylogenetic relationship of P. c-aureum with other nymphalids (see for details). ML analysis exhibited that P. c-aureum formed a monophyletic group with other Nymphalinae species, which were recovered as the sister group to a clade containing the nymphalid subfamilies Apaturinae and Biblidinae with strong support value.
Figure 1. The maximum-likelihood (ML) phylogenetic tree of Polygonia c-aureum and other nymphalid species. Phylogenetic reconstruction was done from a concatenated matrix of 13 protein-coding mitochondrial genes and two ribosomal RNA genes regions in the mitochondrial genome. The numbers beside the nodes are percentages of 1000 bootstrap values (* ≥85%). Alphanumeric terms indicate the GenBank accession numbers.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this paper.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Gan SS, Chen YH, Zuo N, Xia CC, Hao JS. 2014. The complete mitochondrial genome of Tirumala limniace (Lepidoptera: Nymphalidae: Danainae). Mitochondr DNA. 26:1–3.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Hao JS, Sun ME, Shi QH, Sun XY, Shao LL, Yang Q. 2013. Complete mitogenomes of Euploea muciber (Nymphalidae: Danainae) and Libythea celtis (Nymphalidae: Libytheinae) and their phylogenetic implications. ISRN Genomics. 2013:1–14.
- Harvey DJ. 1991. Higher classification of the Nymphalidae, Appen-dix B. In: Nijhout HF, editor. Higher classification of the Nymphalidae, Appendix B. Washington (DC): Smithsonian Institution Press; p. 255–273.
- Kim MJ, WX, Kim KG, Hwang JS, Kim I. 2010. Complete nucleotide sequence and organization of the mitogenome of endangered Eumenis autonoe (Lepidoptera: Nymphalidae). Afr J Biotechnol. 9:735–754.
- McCullagh BS, Marcus JM. 2015. The complete mitochondrional genome of lemon pansy, Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae). J Asia Pac Entomol. 18:749–755.
- Salvato P, Simonato M, Battisti A, Negrisolo E. 2008. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genomics. 9:331.
- Shi QH, Sun XY, Wang YL, Hao JS, Yang Q. 2015. Morphological characters are compatible with mitogenomic data in resolving the phylogeny of nymphalid butterflies (Lepidoptera: Papilionoidea: Nymphalidae). PLoS One. 10:e0124349.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Timmermans MJTN, Lees DC, Thompson MJ, Sáfián S, Brattström O. 2016. Mitogenomics of 'Old World Acraea' butterflies reveals a highly divergent 'Bematistes'. Mol Phylogenet Evol. 97:233–241.
- Wahlberg N, Leneveu J, Kodandaramaiah U, Peña C, Nylin S, Freitas AVL, Brower AVZ. 2009. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc R Soc B. 276:4295–4302.
- Wu LW, Lin LH, Lees DC, Hsu YF. 2014. Mitogenomic sequences effectively recover relationships within brush-footed butterflies (Lepidoptera: Nymphalidae). BMC Genomics. 15:468.
