Abstract
The stick tea thrips, Dendrothrips minowai Priesner (Thysanoptera: Thripidae), is a major pest of tea plantation and poses a considerable economic threat to tea industry. The mitochondrial genome of D. minowai have been sequenced and annotated completely. The entire genome is 14,631 bp in length with an A + T content of 78.53% (GenBank accession No. MF582634). The stick tea thrips mt genome encodes all 37 genes that are typically found in animal mt genomes, consists of 13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes. The gene order is unique and different from that of the other thrips. The A + T-rich region is 149 bp long and contains two poly-T stretchs. Phylogenetic analysis was performed using 13 protein-coding genes with six thrips showed that D. minowai and other five Thripidae species were clustered into a branch, which is formed a sister clade to H. aculeatus (family Phlaeothripidae).
The stick tea thrips, Dendrothrips minowai Priesner (Thysanoptera: Thripidae), is a major pest of tea. Adults and nymphs preferentially feed on tender leaves using rasping–sucking mouth parts, which cause infectious diseases in tea plants and seriously impact tea quality and yield (Bian et al. Citation2016). In this study, the stick tea thrips samples were collected from the tea plantation at Yongchuan, Chongqing, China in 2016, subsequently identified to species by morphology. Voucher specimens (#CQNKY-TH-01-01-01) were deposited at the Insect Collection, Tea Research Institute of Chongqing Academy of Agricultural Science, Chongqing, China.
The complete mitochondrial genome of the stick tea thrips is a typical closed-circular DNA molecule of 14,631 bp in length (GenBank accession No. MF582634). The mt genome of D. minowai is the second smallest mt genome among all sequenced Thysanoptera species and larger than Haplothrips aculeatus (14,616 bp). The total nucleotide composition of the J-strand of the mt genome as follows: A = 40.93% (5988), C = 11.63% (1702), G = 9.84% (1440), and T = 37.60% (5501), with a total A + T content of 78.53%, that is heavily biased toward A and T nucleotides. AT- and GC-skew of the whole J-strand of D. minowai is 0.042 and −0.083, respectively.
The mt genome of D. minowai encodes all 37 genes usually found in animal mt genomes, including 13 protein-coding genes (PCG), two ribosomal RNA genes, and 22 transfer RNA genes. Gene rearrangement occurred frequently among order Thysanoptera, and the Scirtothrips dorsalis South Asia 1 species has a genome consisting of two circular chromosomes (Dickey et al. Citation2015). The mt gene arrangement in D. minowai differs from that of the other thrips. In the mt genome of D. minowai, a total of 29 bp overlaps have been found at eight gene junctions. The mt genome has a total of 309 bp intergenic sequence without the putative A + T-rich region. The intergenic sequences are at 16 locations ranging from 1 to 76 bp, the longest one locates between trnL1 and nad4L. In order Thysanoptera, mt genomes of thrips usually contain two or more putative control regions (Shao et al. Citation2001; Yan et al. Citation2012; Yan et al. Citation2014; Dickey et al. Citation2015). However, there is only one putative control region have been found in the mt genome of D. minowai. The putative control region of this genome is 149 bp long and located between the trnS1 and nad5 genes. The A + T content of this region is 91.95%, the highest level of each region in this mt genome. This region contains two poly-T with 23 bp and 21 bp in length, respectively.
All 22 tRNA genes usually found in the mt genomes of insects have been identified in D. minowai, 18 tRNA genes are encoded by the J-strand and the other four genes are encoded by the N-strand. The nucleotide length of tRNA genes is ranging from 55 bp (trnS1) to 74 bp (trnV), and A + T content is ranging from 66.67% (trnI) to 89.23% (trnS2). Like most insect, 21 tRNA genes have cloverleaf shaped secondary structure and trnS1 gene lacks the dihydrouridine (DHU) arm. The two rRNA genes have been identified on the J-strand in the mt genome: the rrnL gene locates between trnV and trnS2, and the rrnS gene between the trnF and atp8. The length of rrnL and rrnS is 1092 bp and 750 bp, and their A + T content is 79.30% and 80.00%, respectively.
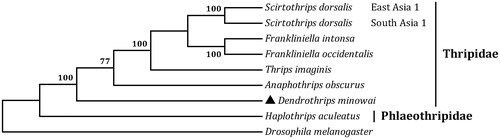
The total length of all 13 protein-coding genes is 10,932 bp, which is accounting for 74.72% of the whole genome sequence. The A + T content of the 13 genes ranges from 71.35% (cox1) to 84.14% (nad6). All of the 13 PCGs start with ATN codons, ATT for atp8, cox1, cob, nad1 and nad4 and ATA for the remainder genes. Two genes, cob and nad2, have incomplete terminal codons consisting of single T nucleotide, and the other PCGs stop with TAA or TAG. The incomplete stop codon T is commonly reported and could produce functional stop codons in polycistronic transcription cleavage and polyadenylation mechanisms (Boore Citation2001). We analyzed the amino acid sequences of 13 PCGs with maximum likelihood (ML) method to learn the phylogenetic relationship of D. minowai with other thrips. The mt genome sequence of Drosophila melanogaster (GenBank accession no. DMU37541) was used as an outgroup. In the tree, D. minowai and other five Thripidae species were clustered into a branch (), which is formed a sister clade to H. aculeatus (family Phlaeothripidae). It infers that the stick tea thrips is closely related to species of Thysanoptera.
Figure 1. The maximum likelihood (ML) phylogenetic tree of Dendrothrips minowai and other moths. The GenBank accession numbers used for tree constructed are as follows: Anaphothrips obscurus (KY498001), Frankliniella intonsa (JQ917403), Frankliniella occidentalis (JN835456), Haplothrips aculeatus (KP198620), Scirtothrips dorsalis East Asia 1 (KM349826), Scirtothrips dorsalis South Asia 1 (KM349827 and KM349828), and Thrips imagines (AF335993).
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bian L, Yang PX, Yao YJ, Luo ZX, Cai XM, Chen ZM. 2016. Effect of trap color, height, and orientation on the capture of yellow and stick tea thrips (Thysanoptera: Thripidae) and nontarget insects in tea gardens. J Econ Entomol. 109:1241–1248.
- Boore JL. 2001. Complete mitochondrial genome sequence of the polychaete annelid Platynereis dumerilii. Mol Biol Evol. 18:1413–1416.
- Dickey AM, Kumar V, Morgan JK, Jara-Cavieres A, Shatters RG Jr, McKenzie CL, Osborne LS. 2015. A novel mitochondrial genome architecture in thrips (Insecta: Thysanoptera): extreme size asymmetry among chromosomes and possible recent control region duplication. Bmc Genomics. 16:439.
- Shao RF, Campbell NJH, Schmidt ER, Barker SC. 2001. Increased rate of gene rearrangement in the mitochondrial genomes of three orders of hemipteroid insects. Mol Biol Evol. 18:1828–1832.
- Yan D, Tang Y, Hu M, Liu F, Zhang D, Fan J. 2014. The mitochondrial genome of Frankliniella intonsa: insights into the evolution of mitochondrial genomes at lower taxonomic levels in Thysanoptera. Genomics. 104:306–312.
- Yan D, Tang Y, Xue X, Wang M, Liu F, Fan J. 2012. The complete mitochondrial genome sequence of the western flower thrips Frankliniella occidentalis (Thysanoptera: Thripidae) contains triplicate putative control regions. Gene. 506:117–124.
