Abstract
The fat sand rats (Psammomys obesus) can easily induce obesity and acquire type 2 diabetes mellitus when they are fed with high-carbohydrate diets. P. obesus is often used as an animal model for studies on diabetes and obesity. We described 16,592 bp of P. obesus mtDNA that contains 13 protein-coding genes (PGCs), two rRNA genes (12S rRNA and 16S rRNA), 22 transfer RNA (tRNA) genes, and one control region (D-loop). The complete mitochondrial genome sequence provided here would be useful for further understanding the evolution of ratite and conservation genetics of P. obesus.
The fat sand rats (Psammomys obesus) belong to the genus Psammomys within the subfamily Gerbillinae of the family Gerbillinae that are widely distributed in North Africa and the Middle East, ranging from Mauritania to the Arabian Peninsula. P. obesus can easily acquire non-insulin-dependent diabetes mellitus and the complications associated with diabetes are (cataracts, pancreatic atrophy, and impaired renal function) from high-caloric foods(Kaiser et al. Citation2005, Citation2012; Ouadda et al. Citation2009).
Apart from previous studies about some biological characters, causes of population depletion, embryonic development, and population genetic diversity (Kaissling et al. Citation1975; FichetCalvet et al. Citation1999; Shenbrot Citation2004; Ouadda et al. Citation2009; Hargreaves et al. Citation2017), molecular studies about P. obesus are limited. No mitochondrial genome of P. obesus is available until now. We will determine the mitochondrial genome of P. obesus in this study. The total genomic DNA was extracted from blood of a male adult P. Obesus in Israel county (31°47'N, 35°13'E) and sequenced with Illumina HiSeq 2000 (San Diego, CA) (Hargreaves et al. Citation2017), the sample of P. Obesus was stored in NCBI (Accession no. SAMN06061930). The complete mitogenome was assembled with MIRA 4.0.2 (San Francisco, CA) (Burlibasa et al. Citation1999) and MITObim 1.9 (Hahn et al. Citation2013). The mitochodrial genome was annotated and drawn by MitoFish 3.30 (http://mitofish.aori.u-tokyo.ac.jp/) ().
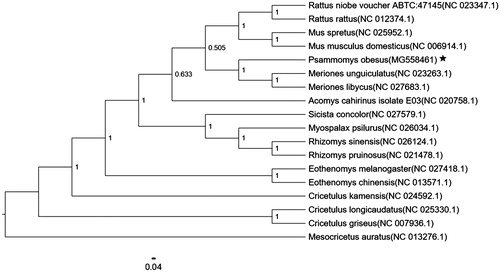
Figure 1. Neighbor-joining molecular phylogenetic tree of 18 species of Glires based on complete mitogenome sequences, with M. auratus as an outgroup. The asterisk indicates the individual sampled in this study. GenBank accession numbers are indicated in brackets.
The complete mitochondrial genome of P. obesus is a double-stranded, circular DNA 16,592 bp in total length (GenBank accession no. MG558461), and includes 13 protein-coding genes, two ribosomal RNA genes (12S rRNA and 16S rRNA), 22 tRNA genes, and one control region (D-loop). The contents of A, G, T, and C are 33.09%, 24.01%, 29.63%, and 12.67%, respectively. GC contents of mitochondrial genome are 36.68%. Twelve of the PCGs use complete (TAA) or incomplete (T-) stop codon. The 12S rRNA and 16S rRNA genes are 951 and 1575 bp, respectively. The lengths of 22 tRNA genes are from 60 bp (tRNA-Ser) to 75 bp (tRNA-Leu). The D-loop length is 1163 bp and lies between the tRNA-Pro and tRNA-Phe genes.
The phylogenetic analysis of 18 mitochondrial genomes using MEGA 7 (Kumar et al. Citation2016) in which Mesocricetus auratus is used as the outgroup indicated that P. obesus and Meriones. unguiculatus are the most closely related species (). The mitogenome of P. obesus would contribute to the understanding of the phylogeny and evolution of Rodentia.
Disclosure statement
The authors report no conflicts of interest, and are alone responsible for the content and writing of the paper.
Additional information
Funding
References
- Burlibasa C, Vasiliu D, Vasiliu M. 1999. Genome sequence assembly using trace signals and additional sequence information. German Conference on Bioinformatics. 99:45–56.
- FichetCalvet E, Jomaa I, Ismail RB, Ashford WR. 1999. Reproduction and abundance of the fat sand rat (Psammomys obesus) in relation to weather conditions in Tunisia. J Zool. 248:15–26.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Hargreaves AD, Zhou L, Christensen J, Marlétaz F, Liu S, Li F, Jansen PG, Spiga E, Hansen MT, Svh P. 2017. Genome sequence of a diabetes-prone rodent reveals a mutation hotspot around the ParaHox gene cluster. Proc Natl Acad Sci USA. 114:7677.
- Kaiser N, Cerasi E, Leibowitz G. 2012. Diet-induced diabetes in the sand rat (Psammomys obesus). Methods Mol Biol. 933:89.
- Kaiser N, Nesher R, Donath MY, Fraenkel M, Behar V, Magnan C, Ktorza A, Cerasi E, Leibowitz G. 2005. Psammomys obesus, a model for environment-gene interactions in type 2 diabetes. Diabetes. 54Suppl 2:S137.
- Kaissling B, De RC, Barrett JM, Kriz W. 1975. The structural organization of the kidney of the desert rodent Psammomys obesus. Anat Embryol. 148:121–143.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870.
- Ouadda A, Levy E, Ziv E, Lalonde G, Sane A, Delvin E, Elchebly M. 2009. Increased hepatic lipogenesis in insulin resistance and Type 2 diabetes is associated with AMPK signalling pathway up-regulation in Psammomys obesus. Biosci Rep. 29:283.
- Shenbrot G. 2004. Habitat selection in a seasonally variable environment: test of the isodar theory with the fat sand rat, Psammomys obesus, in the Negev Desert, Israel. Oikos. 106:359–365.
