Abstract
In this study, the complete mitochondrial genome of the Lasioderma serricorne was sequenced and analysed. The mitochondrial genome is 14,476 bp long and contains 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and two non-coding region. Twenty three genes were found to be encoded by the majority strand and the other 14 genes by minority strand, those similar to that of other insects. The nucleotide compositing of the majority strand is 39.74% of A, 11.20% of C, 40.47% of T, and 10.39% of G. The phylogenetic analysis by maximum-likelihood (ML) method revealed that the L. serricorne was close to Lasioderma redtenbacheri.
The cigarette beetle, Lasioderma serricorne (Fabricius) (Coleoptera: Anobiidae) is a pest of durable grain commodities, spices, and stored tobacco. Larvae cause most feeding damage to commodities (Mahroof Citation2013). However, little information about its genetic characteristic has been reported. Therefore, we determined to sequence the complete mitochondrial genome of L. serricorne using the De Novo sequencing techniques strategy, with the purpose to studying biogeographic, molecular, and population studies. The adult L. serricorne was obtained from tobacco warehouses in Guiyang Tobacco Redrying Factory, Guizhou, China (GPS 26.52094N, 106.67497E), and then was reared on tobacco in lab. The voucher specimens are deposited in Institute of Entomology, Guizhou University, Guiyang, Guizhou, China (GZU-CO-000016). The fourth larvae stage of L. serricorne was washing with 70% ethanol first, and then those were stored in 95% ethanol, the mitochondrial DNA was extracted using De Novo sequencing library, and DNA sequencing at TGS (Total Genomics Solution Institute, Shenzhen, China).
The entire sequence of L. serricorne mitochondrial genome (GenBank accession no. MG592705) is 14,476 bp in length, consisting of 13 protein-coding genes, two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and two non-coding region (one A-T rich region and one control region). Twenty three genes (trnI, trnM, nad2, trnW, cox1, trnL2, cox2, trnK, trnD, atp8, atp6, cox3, trnG, nad3, trnA, trnR, trnN, trnS1, trnE, trnT, nad6, cytB, and trnS2) were found to be encoded by the majority strand (J-strand) and the other 14 genes (trnQ, trnC, trnY, trnF, nad5, trnH, nad4, nad4L, trnP, nad1, trnL1, rrnL, trnV, and rrnS) by minority strand (N-strand), those similar to that of other insects (Lin et al. Citation2017; Singh et al. Citation2017). Overall nucleotide compositions of the majority strand are 39.74% of A, 11.20% of C, 40.47% of T, and 10.39% of G, with an AT content of 80.21%.
The protein-coding genes begin with ATN start codon, such as cox2, nad5, and nad1 start with ATA, atp6, cox3, and nad4 genes employing ATG, while the rest using ATT as a start codon. Three types of stop codon are TAA (cox2, atp6, cox3, nad2, cox1, nad4L, and nad6), TAG (nad5, nad1, atp8, nad3, and cytB), and TTA (nad4). The lrRNA is located between tRNAL1 and A-T rich region, whereas srRNA is accommodated between tRNAV and control region. The 22 tRNA genes vary from 61 to 71 bp in length. The secondary structure of tRNAs exhibited typical clover-leaf structure similar to other insect species. The size of the control region is only 136 bp (37.5% A, 5.15% G, 52.21% T, 5.15% C, with an AT content of 89.71%), and it located between srRNA and tRNAI, and the gene order around the control region is tRNAV-srRNA-control region-tRNAI-tRNAQ-tRNAM, and the order model was reported in some insects (Zhang and Hewitt Citation1997). Another noncodon region is an A-T-rich region, and it located between tRNAV and lrRNA.
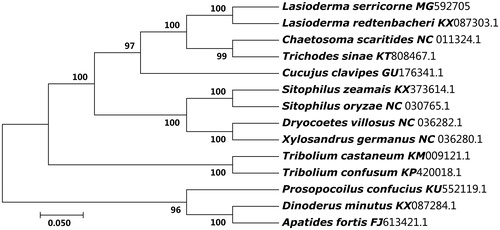
The phylogenetic tree of L. serricorne was constructed with the 13 protein coding genes from 14 Coleoptera beetles by MEGA 7.0 (Kumar et al. Citation2016) using maximum-likelihood (ML) methods (Chen et al. Citation2016). As shown in , the L. serricorne was close to Lasioderma redtenbacheri. Thus, this result supported the monophyly of L. serricorne.
Figure 1. The ML phylogenetic tree of 14 beetles. The nucleotide sequences of the 13 protein-coding genes of each species in the complete or partial (only Lasioderma redtenbacheri) mitochondrial genome were downed from GenBank. The phylogenetic tree was constructed by MEGA 7.0 and Bootstrap support is shown at nodes.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Chen H, Deng S, Yang H, Ma X, Zhu C, Huang H, Li G. 2016. Characterization of the complete mitochondrial genome of Priacanthus tayenus (Perciformes: Priacanthidae) with phylogenetic consideration. Mitochondrial DNA B. 1:243–244.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lin ZQ, Song F, Li T, Wu YY, Wan X. 2017. New mitogenomes of two Chinese stag beetles (Coleoptera, Lucanidae) and their implications for systematics. J Insect Sci (Online). 17:63.
- Mahroof RM. 2013. Stable isotopes and elements as biological markers to determine food resource use pattern by Lasioderma serricorne (Coleoptera: Anobiidae). J Stored Prod Res. 52:100–106.
- Singh D, Kabiraj D, Sharma P, Chetia H, Mosahari PV, Neog K, Bora U. 2017. The mitochondrial genome of Muga silkworm (Antheraea assamensis) and its comparative analysis with other lepidopteran insects. PLoS One. 12:e0188077.
- Zhang D-X, Hewitt GM. 1997. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol. 25:99–120.
