Abstract
Pleurotus cornucopiae is a commercial edible and medicinal fungus. Herein, we determined and analyzed its complete mitochondrial genome. The mitogenome length was 72,134 bp with a GC content of 26.7%, contained 14 conserved protein coding genes, two rRNA genes (rnl and rns), ribosomal protein gene rps3 and 24 tRNA genes. Pleurotus cornucopiae has a similar gene content and gene order of the mitogenome as P. ostreatus and P. eryngii. A phylogenetic analysis based on complete mitogenome in related fungi showed that P. cornucopiae is a member of the order Agaricales, forming a clade with P. ostreatus and P. eryngii, with P. ostreatus as a sister taxa. The mitochondrial genome sequence of P. cornucopiae appeared a promising tool for further studies of the taxonomy and evolution of Pleurotaceae and Agaricales.
The genus Pleurotus, belonging to the family Pleurotaceae (Agaricales, Agaricomycetes, Basidiomycota), is one of the most diverse group of cultivated mushrooms in the world. The Pleurotus species establish a wide range of interactions with other organisms, and are efficient bioconverters of lignocellulosic residues into human food (Tsuneda and Thorn Citation1995; Philippoussis Citation2009). The biodiversity of Pleurotus remains highly investigated and recently, molecular studies have increase our knowledge about intra and inter-specific heterogeneity of the genus Pleurotus using both ribosomal and mitochondrial DNA (Li and Yao Citation2004; Wang et al. Citation2008; Yang et al. Citation2016; Chaudhary and John Citation2017). The edible fungus Pleurotus cornucopiae is emerging as an important species in the genus, with isolation of medicinal metabolites from the fruiting body (Wang et al. Citation2012). The genetic diversity and population structure of P. cornucopiae is well known (Iraçabal and Labarere Citation1994; Zervakis et al. Citation1994; Shnyreva et al. Citation1996; Zhang et al. Citation2007; Zhou et al. Citation2009; Adebayo et al. Citation2016); however, no complete mitogenome is available to date for the species. Here, we report the complete mitogenome of P. cornucopiae (GenBank accession MG652610) to provide new genetic resources and performed a phylogenetic analysis of related taxa of this species.
The strain SWS_15 of Pleurotus cornucopiae is maintained in the Biology Institute, Guangxi Academy of Sciences (Nanning, Guangxi, PR China). Total genomic DNA was extracted as previously described (Xu et al. Citation2017). Library construction and sequencing were processed by Novogene (Beijing, China), according to the Illumina HiSeqX-ten system manufacturer instructions (Illumina, San Diego, CA). The mitogenome of Pleurotus cornucopiae was de novo assembled using ORG.Asm v0.2.05 (https://pythonhosted.org/ORG.asm/) followed by manual curation in Geneious R9 v9.1.6 (Biomatters Ltd, Auckland, New Zealand) as described previously (Hinsinger and Strijk Citation2016; Jiang et al. Citation2016; Xu et al. Citation2017). Genome annotation and phylogenetic analysis were performed with DOGMA (http://dogma.ccbb.utexas.edu/index.html) and Phyml 3.1 (Guindon et al. Citation2010), respectively. Maximum-likelihood (ML) tree was constructed including 10 available mitogenomes of Agaricales and Heterobasidion irregulare as an outgroup ().
Figure 1. ML phylogenetic tree of the 10 available mitogenomes of Agaricales in GenBank, plus the mitogenome of Pleurotus cornucopiae. The tree is rooted with Heterobasidion irregulare. Bootstraps values (1000 replicates) are shown at the nodes. Scale in substitution per site.
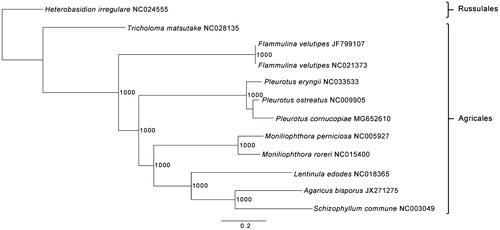
The complete mitogenome of Pleurotus cornucopiae was 72,134 bp in length with a GC content of 26.7%. The mitogenome contained 14 conserved protein coding genes, 2 rRNA genes (rnl and rns), the ribosomal protein gene rps3 and 24 tRNA genes. The 14 conserved protein coding genes respectively encoded the seven ubiquinone reductase subunits of NADH (nad1, nad2, nad3, nad4, nad4L, nad5 and nad6), three cytochrome oxidase subunits (cox1, cox2 and cox3), three ATP synthase subunits (atp6, atp8 and atp9) and the apocytochrome b (cob). The 24 tRNA genes ranged in size from 71 bp to 93 bp, and covered all 20 standard amino acids. Phylogenetic analysis based on ML showed with high support that Pleurotus cornucopiae is a member of Agaricales and grouping with P. ostreatus and P. eryngii, and closely related to P. ostreatus (). This result is consistent to the previous study based on SSU rDNA or LSU rDNA analyses (Gao et al. Citation2008; Chaudhary and John Citation2017), but differs from the result based on ITS data (Shnyreva and Shnyreva Citation2015; He et al. Citation2016). The complete mitochondrial genome of P. cornucopiae provides the community a useful bioresource that will help to delineate taxonomic units in Pleurotus, and will be of interest for further applied researches by selecting the most suitable species for biotechnological and nutritional interest.
Acknowledgements
The authors would like to acknowledge the technical staff of the Biology Institute, Guangxi Academy of Sciences for their assistance in access to the P. cornucopiae strain SWS_15.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Adebayo EA, Oloke JKJ, Azeez MA, Ayandele AA, Majolagbe ON. 2016. Compatibility study using hybridization procedure among Pleurotus genotypes and authentication by enzyme expression and ITSR of rDNA. Russian Agric Sci. 42:423–430.
- Chaudhary MM, John P. 2017. Morphological and molecular characterization of oyster mushroom (Pleurotus cystidiosus). Int J Curr Microbiol Appl Sci. 6:246–250.
- Gao S, Huang CY, Chen Q, Bian YB, Zhang JX. 2008. Phylogenetic relationship of Pleurotus species based on nuclear large subunit ribosomal DNA sequences. J Plant Genet Resour. 9:328–334.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- He XL, Wu B, Li Q, Peng WH, Huang ZQ, Gan BC. 2016. Phylogenetic relationship of two popular edible Pleurotus in China, Bailinggu (P. eryngii var. tuoliensis) and Xingbaogu (P. eryngii), determined by ITS, RPB2 and EF1α sequences. Mol Biol Rep. 43:573–582.
- Hinsinger DD, Strijk JS. 2016. Toward phylogenomics of Lauraceae: the complete chloroplast genome sequence of Litsea glutinosa (Lauraceae), an invasive tree species on Indian and Pacific Ocean islands. Plant Gene. 9:71–79.
- Iraçabal B, Labarere J. 1994. Restriction site and length polymorphism of the rDNA unit in the cultivated basidiomycete Pleurotus cornucopiae. Theoret Appl Genet. 88:824–830.
- Jiang GF, Hinsinger DD, Strijk JS. 2016. Comparison of intraspecific, interspecific and intergeneric chloroplast diversity in Cycads. Sci Rep. 6:31473
- Li XL, Yao YJ. 2004. Assembling phylogenetic tree of based on 28S rDNA sequencing. Mycosystema. 23:345–350.
- Philippoussis AN. 2009. Biotechnology for agro-industrial residues utilisation: production of mushrooms using agro-industrial residues as substrates. Netherlands: Springer; p. 163–196.
- Shnyreva AV, Lomov AA, Mednikov BM, Dyakov JT. 1996. Oyster cap fungi (Pleurotus spp.) species and kinds identification using mating and molecular markers. Mikologiya I Fitopatol. 30:37–44.
- Shnyreva AA, Shnyreva AV. 2015. Phylogenetic analysis of Pleurotus species. Russian J Genet. 51:148–157.
- Tsuneda A, Thorn RG. 1995. Interactions of wood decay fungi with other microorganisms, with emphasis on the degradation of cell walls. Can J Bot. 73:1325–1333.
- Wang SJ, Bao L, Han JJ, Wang QX, Yang XL, Wen HA, Guo LD, Li SJ, Zhao F, Liu HW. 2012. Pleurospiroketals A − E, Perhydrobenzannulated 5,5-Spiroketal Sesquiterpenes from the edible mushroom Pleurotus cornucopiae. J Natural Prod. 76:45–50.
- Wang Y, Zeng FY, Hon CC, Zhang YZ, Leung FCC. 2008. The mitochondrial genome of the Basidiomycete fungus Pleurotus ostreatus (oyster mushroom). FEMS Microbiol Lett. 280:34–41.
- Xu LM, Hinsinger DD, Jiang GF. 2017. The complete mitochondrial genome of the Agrocybe aegerita, an edible mushroom. Mitochondrial DNA Part A. 2:791–792.
- Yang RH, Li Y, Li CH, Xu JP, Bao DP. 2016. The complete mitochondrial genome of the Basidiomycete edible fungus Pleurotus eryngii. Mitochondrial DNA Part B. 1:772–774.
- Zervakis G, Sourdis J, Balis C. 1994. Genetic variability and systematics of eleven Pleurotus species based on isozyme analysis. Mycol Res. 98:329–341.
- Zhang JX, Huang CY, Guan GP, Li HP, Zhang RY, Hu QX. 2007. Inter simple sequence repeat analysis for Pleurotus cornucopiae. Mycosystema. 26:115–121.
- Zhou J, Guo Y, Jia DH, Tan W, Zhang XP, Zheng LY, Huang ZQ. 2009. Hybrid strains of Pleurotus cornucopiae identified by ISSR. Southwest China J Agric Sci. 22:1694–1698.
