Abstract
In this study, the complete mitochondrial genome (mitogenome) of Paragabara curvicornuta was sequenced for the first time by traditional PCR amplification and primer walking methods. As a circular DNA molecule, the entire mitogenome is 15,532 bp in length (GeneBank accession no. KT362742), and consists of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and an AT-rich region. The nucleotide composition is A (39.9%), C (12.0%), G (7.6%), and T (40.4%). Based onthe amino acid sequences of 13 PCGs from 43 noctuoid species as ingroups and two drepanoid species as outgroups, the phylogenetic trees was constructed. The phylogenetic relationships of the families among Noctuoidea are (Notodontidae, (Erebidae, (Nolidae, Noctuidae)). However, the phylogenetic position of P. curvicornuta is still unclear in the family Erebidae that is a monophyletic clade strongly supported by the bootstrap value of 100%.
As a Hypeninae species, Paragabara curvicornuta Kononenko, Han & Matov, Citation2010 is widely distributed in Russian Far East, Japan, Korea and Northeastern China (Kononenko et al. Citation2010). Its phylogenetic relationship among the superfamily Noctuoidea is debated (Zahiri et al. Citation2011; Mitter et al. Citation2017). In July 2014, the P. curvicornuta larvae on Glycine max (Linn.) Merr. were collected from the experimental fields in the campus of Hunan Agricultural University (28° 11'03″N, 113°04'26″E, elevation of 47 m), and the specimens of all the different stages (eggs, larvae, pupae and adults) were obtained. The whole genomic DNA was extracted from a pupa by the standard phenol-chloroform extraction procedure. All the specimens and the genomic DNA were deposited in Insect Museum of Hunan Agricultural University, Changsha City, Hunan Province, China.
The complete mitochondrial genome (mitogenome) of P. curvicornuta was sequenced for the first time by primer walking methods with the reported primers (Gu et al. Citation2016), and compared with the mitogenomes of noctuoid species sequenced. The sequences of P. curvicornuta were proof-read and assembled using the program Geneious 8.12 (Kearse et al. Citation2012). The mitogenome was annotated using the ORF finder (https://www.ncbi.nlm.nih.gov/orffinder) and the nucleotide blasts (https://blast.ncbi.nlm.nih.gov/Blast.cgi) on the NCBI database. To construct the phylogenetic relationship within the superfamily Noctuoidea, the complete mitogenomes of 42 noctuoid species, which plus P. curvitornuta as ingroups, and 2 drepanoid species as outgroups were downloaded. The maximum likelihood (ML) was performed via the software IQTree 1.5.5 (http://www.iqtree.org/) (Lam-Tung et al. Citation2015) with the Number of Bootstrap Replications as 1000.
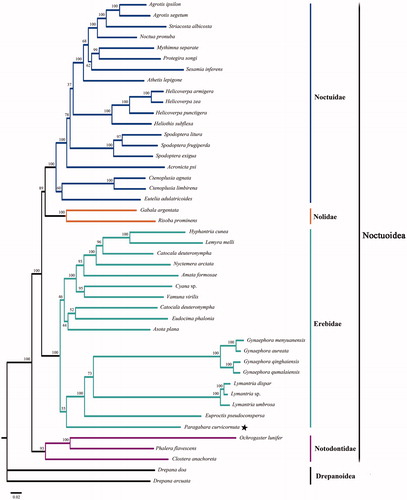
The complete mitogenome of P. curvicornuta is a circular DNA molecule of 15,532 bp in length (GeneBank accession no. KT362742), and consists of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and an AT-rich region. In which, 23 genes are transcribed on the J strand and the remaining 14 are transcribed on the N strand. The nucleotide composition is A (39.9%), C (12.0%), G (7.6%), and T (40.4%), and the AT nucleotide content is 80.4%. There were 273 bp intergenic nucleotides that were dispersed in 16 pairs of neighbouring genes with their length varying from 1 to 52 bp. The length of the A + T-rich region that was located between rrnS and the trnM was 364 bp. The phylogenetic tree based on the putative amino acids sequences from all 13 PCGs with excluding the stop codons is shown in . The relationship within this superfamily Noctuoidea that is a monophyletic clade supported by the bootstrap value of 100% is (Notodontidae, (Erebidae, (Nolidae, Noctuidae)), which is consistent with previously reported results (Regier et al. Citation2017). Furthermore, the family Erebidae as a monophyletic clade is strongly supported by the bootstrap value of 100%. But, the phylogenetic position of P. curvicornuta is still unclear in the family Erebidae, and more data are necessary for further study.
Figure 1. Maximum-likelihood phylogram constructed using 13 PCGs of mitogenomes. Numbers above each node indicates the ML bootstrap support values. All the species’ accession numbers in this study are listed as below: Acronicta psi KJ508060, Agrotis ipsilon NC_022185, Agrotis segetum NC_022689, Amata formosae NC_021416, Asota plana KJ173908, Athetis lepigone NC_036057, Callimorpha dominula NC_027094, Catocala deuteronympha KJ432280, Clostera anachoreta NC_034740, Ctenoplusia agnate NC_021410, Ctenoplusia limbirena NC_025760, Cyana sp KM244679, Drepana arcuata KJ508053, Drepana doa KJ508058, Eudocima phalonia NC_032382, Euproctis pseudoconspersa NC_027145, Eutelia adulatricoides NC_026840, Gabala argentata NC_026842, Gynaephora aureata NC_029162, Gynaephora menyuanensis NC_020342, Gynaephora qinghaiensis NC_029163, Gynaephora qumalaiensis NC_029164, Helicoverpa armigera NC_014668, Helicoverpa punctigera NC_023791, Helicoverpa zea NC_030370, Heliothis subflexa NC_028539, Hyphantria cunea NC_014058, Lemyra melli NC_026692, Lymantria dispar NC_012893, Lymantria sp. KY923068, Lymantria umbrosa NC_035627, Mythimna separate NC_023118, Noctua pronuba KJ508057, Nyctemera arctata KM244681, Ochrogaster lunifer NC_011128, Phalera flavescens NC_016067, Protegira songi NC_034938, Risoba prominens NC_026841, Sesamia inferens NC_015835, Spodoptera exigua NC_019622, Spodoptera frugiperda NC_027836, Spodoptera litura NC_022676, Striacosta albicosta NC_025774, Vamuna virilis NC_026844.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Gu XS, Ma L, Wang X, Huang GH. 2016. Analysis on the complete mitochondrial genome of Andraca theae (Lepidoptera: Bombycoidea). J Insect Sci. 16:105.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kononenko VS, Han H, Мatov AY. 2010. A review of the Eastern Palaearctic genera Paragona Staudinger, 1892 and Paragabara Hampson, 1926 with description of two new species and a new genus (Lepidoptera, Noctuidae: Aventiinae, Hypeninae). Zootaxa. 2679:51–68.
- Lam-Tung N, Schmidt HA, Arndt VH, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating Maximum-Likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Mitter C, Davis DR, Cummings MP. 2017. Phylogeny and evolution of Lepidoptera. Annu Rev Entomol. 62:265–283.
- Regier JC, Mitter C, Mitter K, Cummings MP, Bazinet A, Hallwachs W, Janzen DH, Zwick A. 2017. Further progress on the phylogeny of Noctuoidea (Insecta: Lepidoptera) using an expanded gene sample. Syst Entomol. 42: 82–93.
- Zahiri R, Kitching IJ, Lafontaine JD, Mutanan M, Kaila L, Holloway D, Wahlber N. 2011. A new molecular phylogeny offers hope for a stable family level classification of the Noctuoidea (Lepidoptera). Zool Scr. 40: 158–173.
