Abstract
Herein, the complete Gracilaria edulis mitogenome was determined and analysed. The complete G. edulis mitogenome length was 25,708 bp. The mitogenome contains 50 genes, including 24 protein-coding, two rRNA, and 23 tRNA genes and one unidentified open reading frame (ORF). Of the 24 protein-coding genes, 23 (95.83%) ended with the TAA stop codon, and one (4.17%) with TAG (rpl20). All protein-coding genes in G. edulis were concluded to use the start codon ATG. Mitogenome phylogenetic analysis revealed that G. edulis clustered together with G. changii and G. salicornia. The complete mitogenome sequence provided herein would help understand Gracilaria evolution.
Keywords:
Gracilaria edulis (Rhodophyta) is a marine red alga belonging to the family Gracilariaceae. Gracilaria edulis is a rich source of sulphated polysaccharides, carbohydrates, proteins, vitamins, minerals, and dietary fibres (Sakthivel and Pandima Devi Citation2015). The polyunsaturated esters in G. edulis showed antibacterial activity against several human pathogens. G. edulis extracts can inhibit carcinoma cell activity (Sakthivel et al. Citation2016). To date, studies have mainly focused on the identification of antitumor active compounds in G. edulis (Patra and Muthuraman Citation2013). However, information on its genetics and systematics is limited.
Herein, we determined the complete G. edulis mitogenome sequence. Genomic DNA from one G. edulis individual collected from a population in eastern China (Yingge Sea, Hainan Province, 18°30′36′′N, 108°42′15′′E) was used for genome sequencing. The specimen (GenBank accession number: 2017060064) was deposited at the Culture Collection of Seaweed at the Ocean University of China. Paired-end reads were sequenced using Illumina HiSeq × Ten system (Illumina, San Diego, CA). Approximately, 9 Gb of paired-end (150 bp) sequence data was randomly extracted from the total sequencing output, as input to NOVOPlasty for assembling the mitogenome (Dierckxsens et al. Citation2017). Gracilaria salicornia (GenBank accession number: NC_023784) was used as the seed sequence. tRNA genes were identified using tRNAscan-SE Search Server (Schattner et al. Citation2005). Other mitogenomic regions were annotated from G. salicornia mitogenome using Geneious R10 (Biomatters Ltd., Auckland, New Zealand). Phylogenetic mitogenome analysis was conducted. Bayesian inference (BI) was performed using MrBayes v. 3. 1.2 (Huelsenbeck and Ronquist Citation2001). The phylogenetic analysis was done using two independent runs with four Monte-Carlo Markov Chains running for 1,000,000 generations. Output trees were sampled every 100 generations. The phylogenetic analysis was run until the average standard deviation of split frequencies was below 0.01 and the first 25% of samples was removed as burn-in. Rhodymenia pseudopalmata (KC875852) served as the out-group.
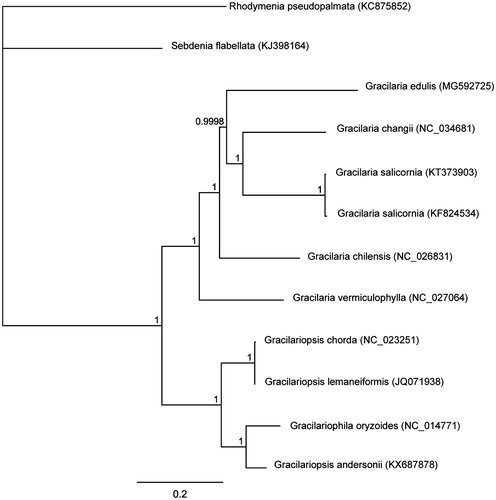
The complete G. edulis (MG592725) mitogenome comprises a circular DNA molecule measuring 25,708 bp in length. The overall A + T content of the complete mitogenome is 75.6%. The mitogenome contains 50 genes, including 24 protein-coding, two rRNA, and 23 tRNA genes and one unidentified open reading frame (ORF). Of the 24 (95.83%) protein-coding genes, 23 ended with the TAA stop codon, and one (4.17%) with TAG (rpl20). All protein-coding genes in G. edulis were concluded to use the start codon ATG. The lengths of two rRNA genes are 2642 bp (rnl rRNA) and 1401 bp (rns rRNA). Bayesian inference showed that G. edulis clustered together with G. changii and G. salicornia (). The complete mitogenome sequence provided herein would help understand Gracilaria evolution.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics (Oxford, England). 17(8):754–755.
- Patra S, Muthuraman MS. 2013. Gracilaria edulis extract induces apoptosis and inhibits tumor in Ehrlich ascites tumor cells in vivo. BMC Complement Altern Med. 13:331.
- Sakthivel R, Muniasamy S, Archunan G, Devi KP. 2016. Gracilaria edulis exhibit antiproliferative activity against human lung adenocarcinoma cell line A549 without causing adverse toxic effect in vitro and in vivo. Food Funct. 7(2):1155–1165.
- Sakthivel R, Pandima Devi K. 2015. Evaluation of physicochemical properties, proximate and nutritional composition of Gracilaria edulis collected from Palk Bay. Food Chem. 174:68–74.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(suppl_2):W686–W689.