Abstract
Paraescarpia echinospica is a conspicuous annelid living in the cold seeps and hydrothermal vents of the Western Pacific region and relying on their endosymbiont bacteria as a source of energy and organic carbon. We report the complete mitochondrial genome of P. echinospica, which is 15,280 bp in length, containing 13 protein-coding genes, two ribosomal RNA genes, 22 tRNA genes and a putative control region. The overall base composition is AT-biased. The control region contains repeated nucleotide motifs. Phylogenetic analyses of the concatenated mitochondrial genes strongly support a sister relationship of P. echinospica with a clade containing Escarpia and Seepiophila.
Paraescarpia echinospica Southward et al., Citation2002 (Read and Fauchald Citation2017) belongs to the deep-sea tubeworm family Siboglinidae. It lacks a digestive tract but relies on the symbiotic chemoautotrophic bacteria harboured in its internal organ called ‘trophosome’ for nutrition (Rouse Citation2001; Bright and Lallier Citation2010). The species commonly occurs in the methane seeps of Papua New Guinea and the Nankai Trough, as well as the hydrothermal vents of the Okinawa Trough (Watanabe et al. Citation2010). Little is known about its phylogenetic relationship with other siboglinids and population connectivity. Here we sequenced the mitogenome of P. echinospica and explored its phylogenetic position in Siboglinidae.
A specimen of P. echinospica was collected from the Haima cold seep located at the northwestern slope of the South China Sea, using the remotely operated underwater vehicle (ROV) Haima in March 2016 (Liang et al. Citation2017), and preserved at −80 °C in the laboratory under the registration number HKUST-QIANT01. Total genomic DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen, Halden, Germany) and used for whole-genome sequencing on an Illumina Hiseq (2 × 150 bp Pair-end reads). Approximately 30 Gb sequence data were assembled de novo using SPAdes v3.9.1 (Bankevich et al. Citation2012) and the contig of mitogenome was verified by BLASTN (Altschul et al. Citation1997) using the mitogenome sequence of Escarpia spicata (Li et al. Citation2015) as the query sequence. Gene annotation was performed with MITOS web server (Bernt et al. Citation2013). The sequence has been deposited in GenBank under accession number MG462707. A maximum likelihood (ML) tree was constructed using the IQ-TREE web service (Trifinopoulos et al. Citation2016).
The circular mitogenome of P. echinospica is 15,280 bp in size, with an overall base composition of 30.11% for A, 22.77% for C, 12.91% for G and 34.21% for T. The genome exhibits codon biases, with an AT content of 63.15% in protein-coding genes. The mitochondrial genome contains 13 protein-coding genes, two ribosomal RNA genes and 22 tRNA genes. The gene order is identical to that of reported siboglinids (Jennings and Halanych Citation2005; Li et al. Citation2015). ATG is the start codon for all genes. Most genes use either TAA or TAG as the stop codon except four genes (nad2, cox1, nad6 and cob) which use a single T as the stop codon. A 593 bp control region lies between trnR and trnH. The control region contains two types of simple repetitive motifs: (TA)n has been found in all reported mitogenomes of siboglinids (Li et al. Citation2015), whereas (ATATATGTGT)n is unique to P. echinospica.
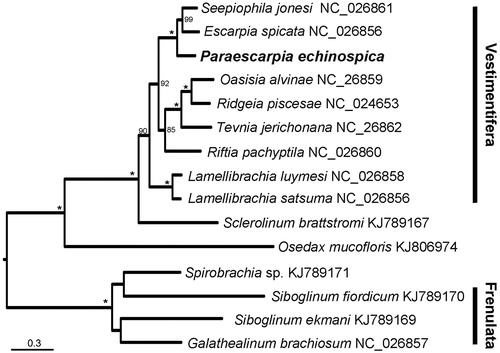
The phylogenetic analysis based on P. echinospica and mitogenetic sequences of all 14 species of siboglinids uploaded on GenBank indicates that Paraescarpia echinospica is sister to a clade comprised of the cold seep siboglinids Seepiophila jonesi and E. spicata (). Based on these data, primers for individual genes can be designed to study the population connectivity, which will help the conservation of these deep-sea animals that are facing increasing human activities such as trawling and mineral extraction (Mengerink et al. Citation2014).
Figure 1. The maximum likelihood (ML) tree of 15 species of Siboglinidae based on the concatenated nucleotide sequences of 13 mitochondrial protein-coding and two ribosomal RNA genes. The number at each node is the bootstrap support value. Asterisks indicate bootstrap support value =100. The number after species name is the GenBank accession number.
Ethical approval
This article does not contain any studies with human subjects by any of the authors.
Acknowledgements
We thank the captain and crew of the research vessel Haiyang VI and the operation team of Haima for collecting the sample.
Disclosure statement
All authors declare that they have no conflict of interest.
Additional information
Funding
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comp. Biol. 19:455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bright M, Lallier FH. 2010. The biology of vestimentiferan tubeworms. Oceanogr Mar Biol Annu Rev. 48:213–266.
- Jennings RM, Halanych KM. 2005. Mitochondrial genomes of Clymenella torquata (Maldanidae) and Riftia pachyptila (Siboglinidae): evidence for conserved gene order in Annelida. Mol Biol Evol. 22:210–222.
- Liang Q, Hu Y, Feng D, Peckmann J, Chen L, Yang S, Liang J, Tao J, Chen D. 2017. Authigenic carbonates from two newly discovered active cold seeps on the northwestern slope of the South China Sea: constraints on fluids sources, formation environments and seepage dynamics. Deep Sea Res Part I. 124:31–41.
- Li Y, Kocot KM, Schander C, Santos SR, Thornhill DJ, Halanych KM. 2015. Mitogenomics reveals phylogeny and repeated motifs in control regions of the deep-sea family Siboglinidae (Annelida). Mol Phylogenet Evol. 85:221–229.
- Mengerink KJ, Van Dover CL, Ardron J, Baker M, Escobar-Briones E, Gjerde K, Levin LA. 2014. A call for deep-ocean stewardship. Science. 344:696–698.
- Read G, Fauchald K. 2017. World Polychaeta database. World Register of Marine Species. Available from: http://marinespecies.org/aphia.php?p=taxdetails&id=328184
- Rouse GW. 2001. A cladistic analysis of Siboglinidae Caullery, 1914 (Polychaeta, Annelida): formerly the phyla Pogonophora and Vestimentifera. Zool J Linnean Soc. 132:55–80.
- Southward EC, Schulze A, Tunnicliffe V. 2002. Vestimentiferans (Pogonophora) in the Pacific and Indian Oceans: A new genus from Lihir Island (Papua New Guinea) and the Java Trench, with the first report of Arcovestia ivanovi from the North Fiji Basin. J Nat Hist. 36:1179–1197.
- Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44:W232–W235.
- Watanabe H, Fujikura K, Kojima S, Miyazaki J, Fujiwara Y. 2010. Japan: vents and seeps in close proximity. In: Kiel S, editor. The vent and seep biota: aspects from microbes to ecosystems. Heidelberg: Springer; p. 379–402.
