Abstract
Asarum sieboldii is a medicinal plant belonging to the Aristolochiaceae family. In this study, complete chloroplast genome sequence of A. sieboldii was characterized through de novo assembly with next generation sequencing data. The chloroplast genome is 193,356 bp long and has the stereotypical tripartite organization consisting of large single copy region and a pair of inverted repeats. The genome contains 78 protein-coding genes, 30 rRNA genes, and 4 tRNA genes. Phylogenetic analysis revealed that A. sieboldii has close relationship with Piper coenoclatum (Piperaceae, Piperales).
The genus Asarum L. (Aristolochiaceae), which consists of approximately 100 species, is usually distributed in temperate regions of Northern Hemisphere, with the center of diversity in Eastern Asia (Oh Citation2007). Since ancient times, the dried root of Asarum species is a widely used drug in traditional medical practices in China, Korea, and Japan (Yamaki et al. Citation1996). In particular, A. sieboldii called “Seshin” in Korean, are used as remedies for aphthous stomatitis, toothache, gingivitis in Korea and China (Zhou Citation1993). Although DNA barcode regions (ITS, rbcL, and matK) of Korean Asarum species have been sequenced and analyzed for species identification and discrimination in genus Asarum (National Institute of Biological Resources (NIBR) Citation2012), the criteria for delimiting the species remain still unclear.
In this study, we determined the chloroplast genome of A. sieboldii to contribute to the classification and development of DNA markers for authentication of Asarum species. The specimen was collected from Mt Deogyu, South Korea (35°51′46.5″ N, 127°44′40.2″ E) and deposited at NIBR herbarium (KB) with the accession number NIBR-VP0000640510. Genome was sequenced using the Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA) and high-quality paired-end reads of ca. 1.7 Gb were used to assemble chloroplast genome (GenBank Accession no. MG551543), as described previously (Kim et al. Citation2015).
The chloroplast genome was 193,356 bp in length with 36.2% overall GC content. Except the chloroplast genome of Pelargonium × hortorum (Chumley et al. Citation2006), the chloroplast of A. sieboldii is the largest terrestrial plant chloroplast genome reported to date. In addition, the genome of A. sieboldii has the stereotypical chloroplast tripartite organization featuring two copies (IRa and IRb) of a 48,401bp IR separating an LSC region of 96,554bp. The SSC region of A. sieboldii is reverse-tandemly duplicated and then integrated into two IRs (Supplementary Material 1–3; https://species.nibr.go.kr/gi/search/chloroplastView/WCN0005156.do). The A. sieboldii genome contains 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The total number of identified genes encoded is 112 genes, with 12 genes duplicated within the SSC integrated into IRs (Supplementary Material 2).
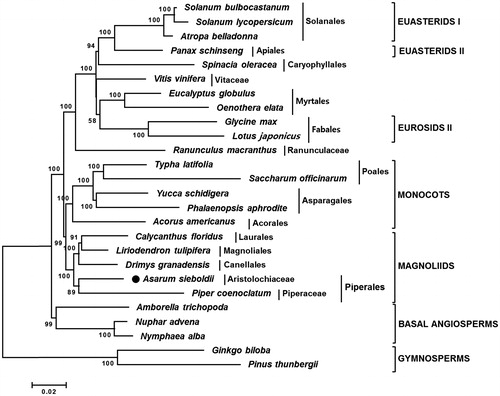
To understand phylogenetic relationship of A. sieboldii with other taxa, a neighbor-joining (NJ) tree was constructed using 62 common protein-coding genes of A. sieboldii and 25 vascular plant taxa (). The NJ tree showed that A. sieboldii was grouped with Piper coenoclatum belonging to the Piperales order (). Chloroplast genome comparison revealed that A. sieboldii showed 62.9% similarity with P. coenoclatum and had 32,732bp larger genome than P. coenoclatum because of extended SC and IRs (Supplementary Materials 1–3).
Figure 1. Neighbor-joining (NJ) tree based on the chloroplast protein-coding genes of 26 taxa including A. sieboldii. Sequences of 65 chloroplast protein-coding gene from 26 taxa were aligned using MAFFT (http://mafft.cbrc.jp/alignment/server/index.html) and used to generate NJ phylogenetic tree by MEGA 6.0 (Tamura et al. Citation2013). The numbers in the nodes indicated the bootstrap support values (>50%) from 1000 replicates. Chloroplast genome sequences used for this tree are: Acorus americanus, NC_010093; Amborella trichopoda, NC_005086; A. sieboldii, MG551543; Atropa belladonna, NC_004561; Calycanthus floridus, NC_004993; Drimys granatensis, DQ887676; Eucalyptus globulus, NC_008115; Ginkgo biloba, NC_016986 (outgroup); Glycine max, NC_007942; Liriodendron tulipifera, NC_008326; Lotus japonicus, NC_002694; Nuphar advena, NC_008788; Nymphaea alba, NC_006050; Oenothera elata, NC_002693; Panax schinseng, NC_006290; Phalaenopsis aphrodite, NC_007499; Pinus thunbergii, NC_001631 (outgroup); Piper coenoclatum, DQ887677; Ranunculus macranthus, NC_008796; Saccharum officinarum, NC_006084; Solanum bulbocastanum, NC_007943; Solanum lycopersicum, DQ347959; Spinacia oleracea, NC_002202; Typha latifolia, NC_013823; Vitis vinifera, NC_007957; Yucca schidigera, NC_032714.
Chae_Eun_Lim_et_al_supplemental_content.zip
Download Zip (564.3 KB)Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, Jansen RK. 2006. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 23:2175–2190.
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655.
- National Institute of Biological Resources (NIBR). 2012. DNA barcode system for Korean indigenous species. Incheon (Republic of Korea): Dongjin Press.
- Oh BU. 2007. Asarum. In: Park CW, editor. The genera of vascular plants of Korea. Seoul: Academy Publishing Company; p.154–155.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Yamaki K, Terabayashi S, Okada M, Park JH. 1996. A new species and a new variety of Asiasaum (Arisolochiaceae) from Korea. J Jpn Bot. 71:1–10.
- Zhou RH. 1993. Resource science of chinese medicinal materials. Beijing: China Medical & Pharmaceutical Sciences Press; p. 202–211.
