Abstract
Lilium washingtonianum Kellogg is native to Western America and grows on slope in high subalpine. In the present study, the complete plastome of L. washingtonianum was sequenced. The plastome sequence was 151,967 bp long, with a large single copy region of 81,393 bp, a small single copy region of 17,352 bp, and two inverted repeat regions of 26,611 bp each. Total 133 genes were identified, including 83 coding genes, 8 ribosomal RNAs, 38 transfer RNAs, and 4 pseudogenes. Among four pseudogenes, pseudo ndhF gene was the first to report among Lilium species so far. The phylogenetic position of L. washingtonianum was sister to L. superbum but American lilies were not monophyletic.
Keywords:
The genus Lilium L. consists of approximately 100 species and the main speciation seems to have occurred after the main landmasses were separated (Pelkonen and Pirttila Citation2012). In terms of many morphological characteristics, the classification of lilies has been modified for long time (Baker Citation1871; Wilson Citation1925; Comber Citation1949; De Jong Citation1974;) but is still controversial because of the discordance with phylogenetic analysis using molecular markers (Hayashi and Kawano Citation2000; Du et al. Citation2014).
Lilium washingtonianum Kellogg is native to Western America and grows on slope in high subalpine (McRae et al. Citation1998). According to Comber (Citation1949), American lilies fell into section Pseudolirion but a clade of American lilies was not strongly supported (Nishikawa et al. Citation1999; Du et al. Citation2014) or they were paraphyletic (Hayashi and Kawano Citation2000; Lee et al. Citation2011). It seems that these low phylogenetic resolutions were caused by poor informative molecular markers in the phylogenetic studies of lilies and that it is necessary to increase gene numbers to solve the low phylogenetic resolution in genus Lilium (Rokas and Carroll Citation2005).
The complete plastome of L. washingtonianum was sequenced in the present study. Total genomic DNA was extracted from fresh leaves of L. washingtonianum, which grew from a seed using DNeasy plant mini kit (Qiagen, Valencia, CA, USA) following the protocol of the manufacture and it is stored in Kyungpook University. Total DNA was sequenced using HiSeq 2500 instrument (Illumina, San Diego, CA, USA).
Genes in the plastome were annotated using Geneious (Kearse et al. Citation2012) by comparing with the previously reported plastome sequences in Lilium species. The plastome sequences of 21 Lilium and 3 Fritillaria species were downloaded from the NCBI database for phylogenetic analysis. A total of 77 genes were extracted from their plastome sequences and each gene was aligned by MAFFT (Katoh et al. Citation2002). The phylogenetic tree was constructed using RAxML (Stamatakis Citation2014) with GTR + G + I model in the CIPRES Science Gateway (Miller et al. Citation2010).
The plastome sequence of L. wasingtonianum (GenBank accession number MG590100) was 151,967 bp long, with a large single copy region of 81,393 bp, a small single copy region of 17,352 bp, and two inverted repeat regions of 26,611 bp each. Total 133 genes were identified, including 83 coding genes, 8 ribosomal RNAs, 38 transfer RNAs, and 4 pseudogenes. Among four pseudogenes, pseudo ndhF gene was the first to report among Lilium species so far.
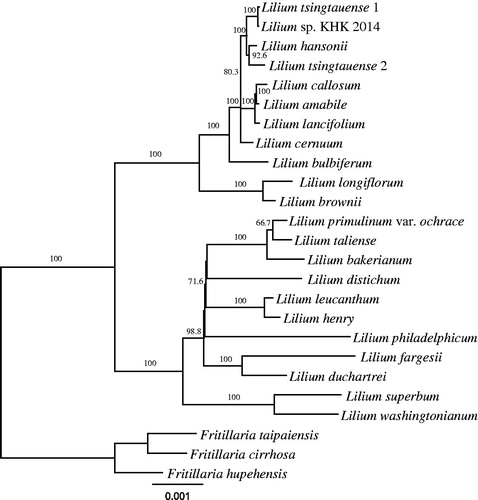
The phylogenetic position of L. washingtonianum was sister to L. superbum but they did not form a clade with L. philadelphicum (), which have belonged to section Pseudolirion as well as L. washingtonianum and L. superbum (Comber Citation1949). Interestingly, L. philadelphicum and L. catesbaei differ from other species in section Pseudolirion by upright flowers and immediate epigeal germination (Pelkonen and Pirttila Citation2012). Our results support that L. philadelphicum should be excluded from the section Pseudolirion in terms of sequence data as well as phenotypic data.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Baker J. 1871. A new synopsis of all the known lilies. Gard Chron. 28:104.
- Comber HF. 1949. A new classification of the genus Lilium. Lily Year Book RHS. 13:86–105.
- De Jong P. 1974. Some notes on the evolution of lilies. Lily Year Book of the North American Lily Society. 27:23–28.
- Du YP, He HB, Wang ZX, Li S, Wei C, Yuan XN, Cui Q, Jia GX. 2014. Molecular phylogeny and genetic variation in the genus Lilium native to China based on the internal transcribed spacer sequences of nuclear ribosomal DNA. J Plant Res. 127:249–263.
- Hayashi K, Kawano S. 2000. Molecular systematics of Lilium and allied genera (Liliaceae): phylogenetic relationships among Lilium and related genera based on the rbcL and matK gene sequence data. Plant Species Biol. 15:73–93.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lee CS, Kim SC, Yeau SH, Lee NS. 2011. Major lineages of the genus lilium (Liliaceae) based on nrDNA ITS sequences, with special emphasis on the Korean species. J Plant Biol. 54:159–171.
- McRae EA, Austin-Mcrae E, MacRae E. 1998. Lilies: a guide for growers and collectors. Timber Press Portland.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); New Orleans, LA.
- Nishikawa T, Okazaki K, Uchino T, Arakawa K, Nagamine T. 1999. A molecular phylogeny of Lilium in the internal transcribed spacer region of nuclear ribosomal DNA. J Mol Evol. 49:238–249.
- Pelkonen V, Pirttila A. 2012. Taxonomy and phylogeny of the genus lilium. Floriculture Ornamental Biotech. 6:1–8.
- Rokas A, Carroll SB. 2005. More genes or more taxa? The relative contribution of gene number and taxon number to phylogenetic accuracy. Mol Biol Evol. 22:1337–1344.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wilson E. 1925. The lilies of Eastern Asia, a monograph. London.