Abstract
Pimpinella rhomboidea var. tenuiloba is an endemic species in China. The complete chloroplast genome sequence of P. rhomboidea var. tenuiloba was generated by de novo assembly using whole genome next generation sequencing. The genome was 146,655 bp in length, composed of four distinct regions such as large single copy (LSC) region of 94,684 bp, small single copy (SSC) region 17,537 bp and a pair of inverted repeat regions of 17,217 bp. The genome annotation predicted a total of 113 genes, including 80 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. Phylogenetic analysis with the reported chloroplast genomes revealed that P. rhomboidea var. tenuiloba has close relationship with genus Angelica.
Keywords:
Pimpinella rhomboidea Diels var. tenuiloba Shan & Pu (Apiaceae, Apioideae), a perennial herb which is endemic to mountainous region of West Sichuan, China, growing at an elevation of 2200–4000 m (Tan et al. Citation2015). Based on our previous work (Tan et al. Citation2015), P. rhomboidea var. tenuiloba was treated as a synonym of Melanosciadium bipinnatum (Shan and Pu) Pimenov & Kljuykov, and has been corroborated to nest in Angelica sensu lato within Selineae (Zhou et al. Citation2009, Liao et al. Citation2013, Wang et al. Citation2014). Here, we assembled the complete chloroplast genome of P. rhomboidea var. tenuiloba, to explore the reasonably systematic position of this taxon.
The mature leaves of P. rhomboidea var. tenuiloba were collected from Mt. Balang (30°53′55.19″N 102°54′38.06″E), Sichuan Province, China. Voucher specimens were deposited in SZ (Sichuan University Herbarium). The isolated genomic DNA was manufactured to average 350bp paired-end (PE) library using the Illumina Hiseq platform, and sequenced by Illumina genome analyzer (Hiseq PE150). Chloroplast genome-related reads were sieved by mapping to all existent chloroplast genomes of Apioideae in GenBank using Bowtie2. Contigs, assembled using SOAPdenovo2 (Luo et al. Citation2012) and SPAdes (Bankevich et al. Citation2012), were sorted and joined into a single-draft sequence using Geneious 11.04 (Kearse et al. Citation2012) by comparison with the chloroplast sequences of Angelica. The draft sequence was then corrected manually by clean reads mapping using bowtie2 (Langmead and Salzberg Citation2012) and Tablet (Milne et al. Citation2013). The genes in chloroplast genome were predicted using Geneious by pairwise align with sequence of Angelica gigas, and revised manually.
The chloroplast genome of P. rhomboidea var. tenuiloba (Genbank accession no. MG719855) was a circular molecular genome with a size of 146,655 bp in length, which was composed of four distinct regions such as large single copy (LSC) region of 94,684 bp, small single copy (SSC) region 17,537 bp and a pair of inverted repeat regions of 17,217 bp. The overall GC content was 37.5%. The chloroplast genome contained 80 protein-coding genes, 29 tRNA genes and 4 rRNA genes.
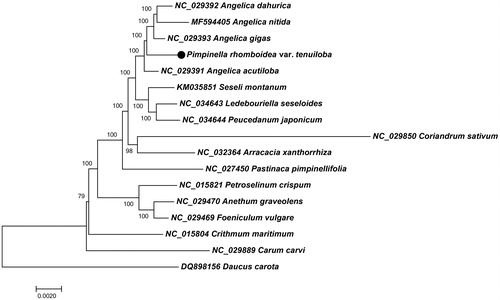
In order to understand the phylogenetic relationship between P. rhomboidea var. tenuiloba and related species, the complete chloroplast genome sequences of 12 genera (16 species) from apioid superclade and one outgroup (Daucus carota) were aligned by MAFFT (Katoh et al. Citation2002) and trimmed properly by trimAl v1.4 (Capella-Gutierrez et al. Citation2009). The evolutionary history was inferred by using the Neighbour-joining method in MEGA7.0 (Kumar et al. Citation2016). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100,000 replicates) are shown next to the branches (). As was expected, P. rhomboidea var. tenuiloba was placed within Angelica species, and occurred as a sister to the clade of A. dahurica, A. nitida and A. gigas with 100% BS value.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25:1972–1973.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28: 1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33:1870–1874.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–U354.
- Liao CY, Downie SR, Li QQ, Yu Y, He XJ. 2013. New insights into the phylogeny of Angelica and its allies (Apiaceae) with emphasis on east Asian species, inferred from nrDNA, cpDNA, and morphological evidence. Syst Bot. 38:266–281.
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 14:193–202.
- Tan JB, Wang ZX, Xie DF, He XJ. 2015. Pimpinella rhomboidea var. tenuiloba is a synonym of Melanosciadium bipinnatum (Apiaceae). Nordic J Bot. 33:659–661.
- Wang ZX, Downie SR, Tan JB, Liao CY, Yu Y, He XJ. 2014. Molecular phylogenetics of Pimpinella and allied genera (Apiaceae), with emphasis on Chinese native species, inferred from nrDNA ITS and cpDNA intron sequence data. Nordic J Bot. 32:642–657.
- Zhou J, Gong X, Downie SR, Peng H. 2009. Towards a more robust molecular phylogeny of Chinese Apiaceae subfamily Apioideae: additional evidence from nrDNA ITS and cpDNA intron (rpl16 and rps16) sequences. Mol Phylogenet Evol. 53:56–68.