Abstract
In this study, the complete mitogenome sequence of Conocephalus melaenus (Orthoptera: Tettigoniidae) has been decoded for the first time by next generation sequencing method. The overall base composition of C. melaenus mitogenome are 37.1% for A, 17.3% for C, 11.1% for G and 34.4% for T, and has high AT content of 71.5%. The complete mitochondrial genome of C. melaenus is 15,923 bp in length, and consisted of 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes and one A + T-rich region. An unusual feature of the C. melaenus mitogenome is the presence of a tRNASer(UCN)-like sequence on the J-strand, and the tRNASer(UCN)-like sequence have proper folding structures and anticodons sequences. Using the 13 PCGs and 2 rRNA of C. melaenus, together with 17 other close-related and two outgroup species, we constructed Bayesian inference phylogenetic tree to verify the accuracy and utility of new determined mitogenome sequences. The complete mitogenome of C. melaenus provides valuable molecular data for further phylogenetic and evolutionary analysis in Tettigoniidae.
Conocephalus melaenus belongs to Conocephalinae, Tettigoniidae, Orthoptera. The specimen of C. melaenus was collected from Xinyang, Henan, China (N 31°48′, E 114°04′) in 9th August 2009, and was now deposited in Molecular and Evolutionary Lab in Shaanxi Normal University. The total DNA was isolated from the leg muscle of the specimen using the phenol-chloroform extraction method (Zhou et al. Citation2010). We sequenced the genome of C. melaenus using the next-generation sequencing technology, the raw reads and bases generated from HiSeq 2500 (Illumina, San Diego, CA) were 11,073,675 and 2.79Gb separately, and the length of one read is about 125 bp. Then the CLC Genomics Workbench 9.0 (CLC Bio, Aarhus, Denmark) was used to remove low-quality reads. Using the Conocephalus maculatus mitochondrial genome as the reference sequence, we assembled the mitochondrial genome of C. melaenus using Mira 4.0.2 and MITObim 1.7 (Hahn et al. Citation2013). The annotation of protein-coding genes (PCGs), rRNA genes and A + T-rich region were conducted using Geneious 9.1.2 (Biomatters Ltd., Auckland, New Zealand). tRNA genes were predicted using online software tRNAScan-SE 1.21 and MITOS (Lowe and Eddy Citation1997; Bernt et al. Citation2013).
The complete mitogenome of C. melaenus is 15,923 bp in length and has been deposited in GenBank (accession no. KY407794). The overall base composition of C. melaenus mitogenome are 37.1% for A, 17.3% for C, 11.1% for G and 34.4% for T, and has high AT content of 71.5%. It comprises 13 PCGs, 22 tRNA genes, two rRNA genes and one A + T-rich region. The orientation and gene order of the C. melaenus mitogenome are identical to the hypothesized ancestral arthropod and the reference C. maculatus. One tRNASer(UCN)-like sequence on the J-strand was found in the C. melaenus mitogenome. The tRNASer(UCN)-like sequence is 68 bp in length and located between tRNASer(UCN) and nad1. The tRNASer(UCN)-like sequence has 94.1% identity, a similar folding structure and the same anticodon sequence with tRNASer(UCN). Additionally, a 356 bp intergenic spacer was found between nad1 and tRNALeu(CUN). 16S rRNA and 12S rRNA genes are 1315 bp and 837 bp in length separately, and the control region is as long as 654 bp, with an AT bias of 79.1%.
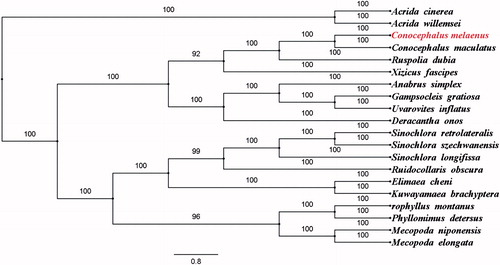
To furthermore validate the new determined sequences and annotations, the concatenated mitogenome dataset (PCGs and rRNAs) of C. melaenus in this study, and together with other 19 species from GenBank, including 17 species from Tettigoniidae and two Caeliferan outgroup species were used to perform phylogenetic analysis (). The phylogenetic tree showed that C. melaenus and C. maculatus were grouped together with a 100% bootstrap support. The complete mitogenome of C. melaenus provides valuable molecular data for further phylogenetic and evolutionary analysis in Tettigoniidae.
Figure 1. The Bayesian inference phylogenetic tree of the Conocephalus melaenus in this study and other 19 species including 17 from Tettigoniidae and two outgroup species based on mitochondrial PCGs and rRNAs concatenated dataset. The GenBank accession numbers for tree construction is listed as follows: Acrida cinerea (GU344100), Acrida willemsei (EU938372), Anabrus simplex (NC_009967), Conocephalus maculatus (NC_016696), Deracantha onos (NC_011813), Elimaea cheni (NC_014289), Gampsocleis gratiosa (NC_011200), Kuwayamaea brachyptera (NC_028159), Mecopoda elongata (NC_021380), Mecopoda niponensis (NC_021379), Orophyllus montanus (KT345951), Phyllomimus detersus (NC_028158), Ruidocollaris obscura (NC_028160), Ruspolia dubia (NC_009876), Sinochlora longifissa (NC_021424), Sinochlora retrolateralis (KC467056), Sinochlora szechwanensis (KX354724), Uvarovites inflatus (NC_026231) and Xizicus fascipes (NC_018765).
Disclosure statement
The author reports no conflict of interest.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Lowe T, Eddy S. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Zhou Z, Ye H, Huang Y, Shi F. 2010. The phylogeny of Orthoptera inferred from mtDNA and description of Elimaea cheni (Tettigoniidae: Phaneropterinae) mitogenome. J Genet Genomics. 37:315–324.
