Abstract
The complete mitochondrial genome of Thryssa hamiltonii has been determined. The whole sequence was 16,894 bp in length and included 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes and one control region (D-loop). The overall base composition is A 30.77%, C 27.97%, G 16.25%, T 25.01%, with a slightly A + T bias of 55.78%. With the exception of ND6 and eight tRNA genes, all other mitochondrial genes are encoded on the heavy strand. Three tandem repeat sequences were observed in the control region. Phylogenetic tree was constructed based on 13 protein-coding genes sequences of 21 clupeoidei species, and the result showed that T. hamiltonii is most closely related to T. dussumieri. These mitogenome sequence data will be useful for population genetics and phylogenetic analysis of the Clupeoidei.
Thryssa hamiltonii, which belongs to Engraulinae, Engranlidae, Clupeoidei, is distributed in the western Indo-Pacific (PJP et al. Citation1988; J.S. Citation1994). Although there were some studies on its fishery biology (Qin et al. Citation2011) and population structure (AL-HASSA Citation1988), little information about its genetic characteristics is available. In order to find new DNA markers for the future research of population genetics and phylogenetics and taxology, we determined the complete mitogenome of T. hamiltonii (GenBank accession number MF668229) by PCR amplification and primer walking sequence method.
Thryssa hamiltonii was collected from the South China Sea (18°12'30”N 109°28'49”E) and stored in a refrigerator of −80 °C with accession number 20161015TH04. The specimen was identified based on the morphologic features and COI gene. Muscle tissues of individual specimens for molecular analysis were reserved in ethanol absolute. Whole genomic DNA was extracted by using the phenol-chloroform method (Barnett and Larson Citation2012). The quality of the genomic DNA was checked using 1% agarose gel. The universal primers (Ivanova et al. Citation2007) were designed from the conserved regions of the complete mitochondrial genome sequences of 26 Clupeiformes species from GenBank database.
The complete mitochondrial genome of T. hamiltonii was 16,894 bp in length, consisting of 13 protein-coding genes, 22 transfer RNA genes (tRNA), two ribosomal RNA genes (12S rRNA and 16S rRNA) and one control region (D-loop). Except ND6 and eight tRNAs (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, Pro), other genes were encoded on the heavy strand. The mitochondrial base composition is A 30.77%, C 27.97%, G 16.25%, T 25.01%, respectively. The A + T content (55.78%) is higher than G + C content, in common with other Clupeoidei mitogenomes (Bi and Chen Citation2011; Li et al. Citation2012; Qiao et al. Citation2012; Bo et al. Citation2013; Wang et al. Citation2015; Zhang et al. Citation2016). Twelve protein-coding genes start with ATG except COX1 with GTG. For the stop codon, ND6 ends with TAG, seven genes with TAA, ND2, ND3, COX2, ND4 and CYTB with an incomplete TA or T. The 12 S rRNA (954 bp) is located between tRNAPhe and tRNAVal genes, and 16S rRNA (1690 bp) is located between tRNAVal and tRNALeu genes. The control region (D-Loop) typically located between tRNAPro and tRNAPhe genes, is 1246 bp in length. Three tandem repeat sequences were observed in the control region. The shortest motif was 39 bp with five repeats, and other two motifs were both 117 bp with two repeats. The symbolic structures of the control region are observed as in other fishes, such as the TAS-cTAS, central conserved sequence blocks (CSB-F, D, B, A), CSB 2-3, G-box (GTGGGGG), and a pyrimidine tract (Guo et al. Citation2003; Gong et al. Citation2015).
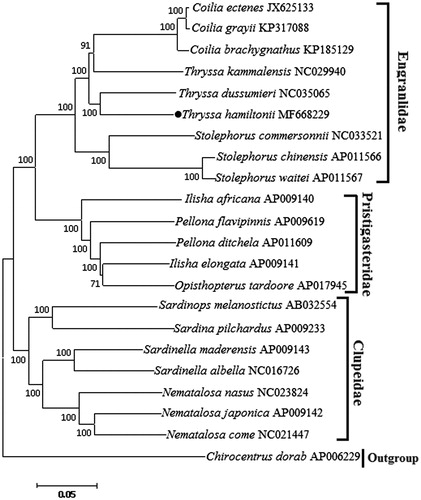
Phylogenetic tree () was constructed using the neighbour-joining (NJ) method based on the 13 protein-coding genes of 21 clupeoidei species. The result shows that T. hamiltonii is most closely related to T. dussumieri. We expect the present results will further facilitate for the study on the taxonomy, population genetic structure and phylogenetic relationships of Clupeoidei.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Al-Hassa LAJ. 1988. Genetic variation and populations structure in Thryssa hamiltonii and Nematalosa nasus in Iraqi waters. J Fish Biol. 33:219–220.
- Barnett R, Larson G. 2012. A phenol-chloroform protocol for extracting DNA from ancient samples. Methods Mol Biol. 840:13–19.
- Bi YH, Chen XW. 2011. Mitochondrial genome of the American shad Alosa sapidissima. Mitochondrial DNA. 22:9–11.
- Bo Z, Xu T, Wang R, Jin X, Sun Y. 2013. Complete mitochondrial genome of the Osbeck's grenadier anchovy Coilia mystus (Clupeiformes, Engraulidae)). Mitochondrial DNA. 24:657–659.
- Gong L, Shi W, Si LZ, Wang ZM, Kong XY. 2015. The complete mitochondrial genome of peacock sole Pardachirus pavoninus (Pleuronectiformes: Soleidae) and comparative analysis of the control region among 13 soles. Mol Biol. 49:408–417.
- Guo X, Liu S, Liu Y. 2003. Comparative analysis of the mitochondrial DNA control region in cyprinids with different ploidy level. Aquaculture. 224:25–38.
- Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN. 2007. Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes. 7:544–548.
- Nelson JS. 1994. Fishes of the world. 3rd ed. Wiley; p. 118–123.
- Li M, Zou K, Chen Z, Chen T. 2012. Mitochondrial genome of the Chinese gizzard shad Clupanodon thrissa (Clupeiformes: Clupeidae) and related phylogenetic analyses. Mitochondrial DNA. 23:438–440.
- PJP W, GJ N, T W. 1988. FAO species catalogue. v. 7: Clupeoid fishes of the world (Suborder Clupeoidei). An annotated and illustrated catalogue of the herrings, sardines, pilchards, sprats, shads, anchovies and wolf-herrings. Part 2 – Engraulidae. FAO Fish Synop. 125:305–579.
- Qiao H, Cheng Q, Chen Y, Chen W, Zhu Y. 2012. The complete mitochondrial genome sequence of Coilia ectenes (Clupeiformes: Engraulidae). Mitochondrial DNA. 24:123–125.
- Qin J, Chen H, Cai W, Yang T, Jia X. 2011. Effect of di-n-butyl phthalate (DBP) on biochemistry indicators of Thryssa hamiltonii. South China Fisheries Sci. 7:29–34.
- Wang S, Wang B, Hu M, Wang F, Wu Z. 2015. The complete mitochondrial genome of Coilia brachygnathus (Clupeiformes: Engraulidae: Coilinae). Mitochondrial DNA Part A. 27:4084–4085.
- Zhang N, Song N, Gao T. 2016. The complete mitochondrial genome of Coilia nasus (Clupeiformes: Engraulidae) from Ariake Sea. Mitochondrial DNA A DNA Mapp Seq Anal. 27:1518–1519.