Abstract
Prunella vulgaris L. is an important medicinal herb widely used in China and western countries. Its natural distribution occurs in various habitats throughout northern hemisphere. In present study, we assembled and characterized its whole chloroplast (cp) genome based on Illumina pair-end sequencing data. The complete chloroplast genome size is 156,132 bp. It contained 134 genes, including 89 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. 8 gene species had two copies. The overall GC content of this genome was 37.9%. A further phylogenomic analysis of Lamiaceae, including 29 taxa, was conducted for the placement of P. vulgaris.
Prunella vulgaris L. (Lamiaceae), is a perennial herb known as ‘common self-heal’ found in temperate to subtropical climates throughout Europe, Asia, and North America. It is a long used herbal medicine in both Europe and China for treatment of fevers, sore mouth, and wound healing (Kim et al. Citation2014; Lou et al. Citation2014; Qu et al. Citation2017). In the present study, we assembled and characterized the complete plastome of P. vulgaris. It will provide potential genetic resources, such as SNPs, DNA barcodes and cpSSR markers, for further population genetics and germplasm diversity conservation studies.
Fresh leaves of P. vulgaris were collected from Zhejiang province, China. A voucher specimen (accession number HYW170001) has been deposited at the University of Macau. The total DNA of P. vulgaris was extracted using CTAB. A short-insert (500 bp) paired-end library was prepared and sequenced using by HiSeqTM 2500 (Illumina, San Diego, CA, USA). Approximately 16 million high quality clean reads were generated. Then the cp genome was aligned, assembled, and annotated by CLC Genomics Workbench v7.5 software (CLC Bio, Aarhus, Denmark), GeSeq (Tillich et al. Citation2017), tRNAscan-SE v1.3.1 (Schattner et al. Citation2005). The rRNAs and protein-coding genes (PCGs) were further validated by comparing with a custom BLAST database of Lamiaceae plastomes annotations.
The complete cp genome of P. vulgaris (GenBank Accession MG589640) was 151,342 bp in size, containing a large single copy region (LSC) of 82,689 bp and a small single copy region (SSC) of 17,428 bp, which were separated by a pair of inverted repeats (IRs) of 25,613 bp. The cp genome contained 134 genes, including 89 protein-coding genes (PCGs), 37 tRNA genes, and 8 rRNA genes. Most of the genes occurred as single-copy, while 20 genes had two copies, which included 9 PCG genes (ndhB, rpl2, rpl23, rps7, rps12, rps19, ycf1, ycf2, and ycf15), 7 tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and all 4 rRNA species (rrn4.5, rrn5, rrn16 and rrn23). Two PCGs (clpP, and ycf3) contained two introns while another 8 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1 and rps16) had one intron each. The overall GC content of the P. vularis cp genome was 37.9%, with the corresponding values of LSC, SSC and IR regions being 36.0, 31.8 and 43.1%, respectively.
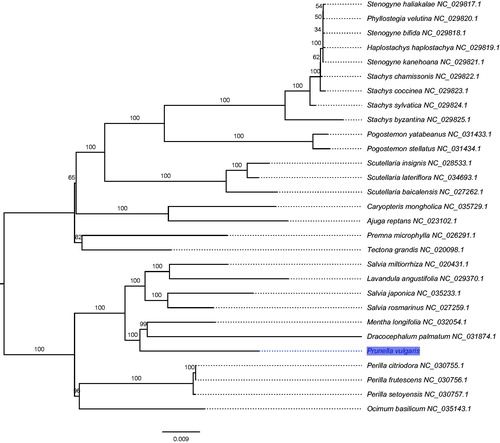
The phylogenetic analyses were conducted based on 86 protein-coding genes of 29 complete cp genomes in Lamiaceae to uncover the phylogenetic placement of P. vulgaris. The maximum likelihood (ML) inference was performed using GTR + Γ model with 1000 bootstrap replicates in RAxML v.8.2.8 (Stamatakis Citation2014). The result revealed that P. vulgaris was clustered with a clade of Dracocephalum + Mentha with 100% bootstrap values (). In conclusion, the complete cpDNA of P. vulgaris is decoded for the first time in this study and provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
References
- Kim HI, Quan FS, Kim JE, Lee NR, Kim HJ, Jo SJ, Lee CM, Jang DS, Inn KS. 2014. Inhibition of estrogen signaling through depletion of estrogen receptor alpha by ursolic acid and betulinic acid from Prunella vulgaris var. lilacina. Biochem Bioph Res Co. 451:282–287.
- Lou H, Zheng S, Li T, Zhang J, Fei Y, Hao X, Liang G, Pan W. 2014. Vulgarisin A, a new diterpenoid with a rare 5/6/4/5 ring skeleton from the Chinese medicinal plant Prunella vulgaris. Org. Lett. 16: 2696–2699.
- Qu Z, Zhang J, Yang H, Gao J, Chen H, Liu C, Gao W. 2017. Prunella vulgaris L., an edible and medicinal plant, attenuates scopolamine-induced memory impairment in rats. J. Agric. Food Chem. 65: 291–328.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbrichtjones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.