Abstract
Vicia sepium L., as a pan-temperate species, it is suitable as a constituent part of grazed grasslands because it can be resistant to winter kill and it contains abundant proteins. In this study, complete chloroplast genome of V. sepium was explored and phylogenetic analysis was done. The result showed that the whole genome was 124,095 bp long with IR loss. The overall GC content was 35.03%. A total of 109 genes were identified, including 80 protein-coding, 30 transfer RNA, and four ribosome RNA genes. Similar to most of the IRLC plastoms, rpl22, rps16 and one intron of clpP were lost. The phylogenetic analysis of protein-coding genes from 25 Fabaceae species belonging to IRLC showed that V. sepium was similar to V. sativa. The chloroplast genome reported here will promote our understanding of the phylogeny and evolution of the genus Vicia.
Vicia sepium L., in subfamily Papilionoideae of Fabaceae, is a well-known species widely distributed in Europe, Central Asia, and North Asia, introduced from the Old World to North America. As a pan-temperate species, it is suitable as a constituent part of grazed grasslands because it can be resistant to winter kill and it contains abundant proteins (Maršalkienė Citation2016). However, the taxonomic history of Vicia L. is confused and contentious which has led misapplication of names of V. sepium. Compared to other angiosperms, the inverted-repeat-lacking clade (IRLC) including V. sepium exists diversity of changes in structural rearrangements of chloroplast genome (Sabir et al. Citation2014). Then, there are disagreements among authors concerning the classification of Vicia L. because of its high morphological variability (Shiran and Raina Citation2001; Iamonico et al. Citation2017). Therefore, we sequenced the complete chloroplast genome of V. sepium to enhance our understanding of its evolution processes and breeding.
The mature leaves of V. sepium were obtained from a natural population in Dongting Lake region (28°48′46.06″N, 112°21′10.19″E) under liquid N2 and stored at −80 °C with accession number 20170707JJ. The total chloroplast DNA was extracted with Plant Chloroplast Purification Kit (BTN120308) and Column Plant DNA Extraction Kit and sequenced by using Illumina Hiseq 4000 Platform. Filtration and annotation of the whole genome and drawing of a circular map referred to Zhang’s method (Zhang et al. Citation2017) by using Trimmomatic v0.32, DOGMA and cpGAVAS, OGDRAW. The complete chloroplast genome was submitted to the NCBI database under the accession number MG682352.
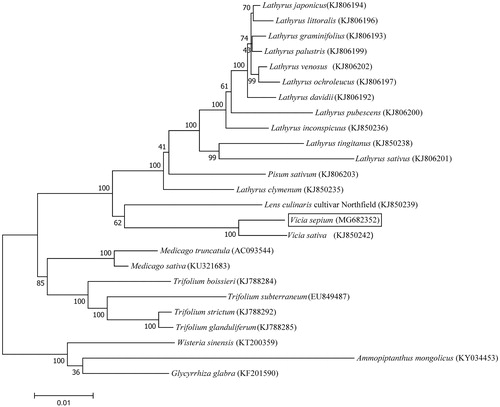
The complete chloroplast genome of V. sepium was 124,095 bp long with IR loss. The overall GC content was 35.03%. A total of 109 genes were identified, including 80 protein-coding, 30 transfer RNA, and four ribosome RNA genes. A total of 12 genes (ndhA, rrn23S, ndhB, ycf2, rpl2, accD, atpF, ropC1, rpoB, clpP, ycf3, psaA) contained introns: ycf3 contained three introns, ycf2 contained two introns and the rest of the genes contained a single intron. Similar to most of the IRLC plastoms, rpl22, rps16 and one intron of clpP were lost. Phylogenetic relationships were presented using 54 conserved chloroplast protein sequences shared among 24 published Fabaceae species and V. sepium by using the maximum likelihood method with 1000 bootstrap replicates based on Muscle progress in MEGA7.0 () (Kumar et al. Citation2016). Our phylogenetic analysis confirmed the relationship between V. sepium and V. sativa within the Fabaceae. The chloroplast genome reported here will promote our understanding of the phylogeny and evolution of the genus Vicia.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Iamonico D, Iberite M, Abbate G. 2017. Vicia incisa (Fabaceae): taxinomical and chorological notes. Ann Bot (Roma). 7:57–65.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33:1870–1874.
- Maršalkienė N. 2016. Flowering spectrum diversity of Vicia sepium. Biologija. 62:116–123.
- Sabir J, Schwarz E, Ellison N, Zhang J, Baeshen NA, Mutwakil M, Jansen R, Ruhlman T. 2014. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol J. 12:743–754.
- Shiran B, Raina SN. 2001. Evidence of rapid evolution and incipient speciation in Vicia sativa species complex based on nuclear and organellar RFLPs and PCR analysis. Genet Resour Crop Evol. 48: 519–532.
- Zhang W, Zhao YL, Yang GY, Tang YC, Xu ZG. 2017. Characterization of the complete chloroplast genome sequence of Camellia oleifera in Hainan, China. Mitochondrial DNA B. 2:843–844.