Abstract
Rosa roxburghii Tratt. is a famous Chinese traditional medicine with long history, here we characterized the whole chloroplast (cp) genome sequence of R. roxburghii by Illumina pair-end sequencing. The complete cp genome was 156,749 bp in length, containing a large single copy (LSC) region of 85,862 bp and a small single copy (SSC) region of 18,791 bp, which were separated by a pair of 26,053 bp inverted repeat regions (IRs). The genome contained 139 genes, including 88 protein-coding genes (82 PCG species), 39 tRNA genes (32 tRNA species), and eight ribosomal RNA genes (four rRNA species). Most of the gene species occur as a single copy, while 17 gene species occur in double copies. The overall AT content of R. roxburghii cp genome is 62.8%, while the corresponding values of the LSC, SSC, and IR regions are 64.8, 68.7, and 57.4%, respectively. Further, phylogenetic analysis suggested that R. roxburghii is closely related to R. odorata var. gigantea.
Introduction
Rosa roxburghii Tratt. is a wild perennial shrubs of Rosaceae that originates from Southwestern China. For hundreds of years, R. roxburghii has been used as traditional medicine in China. With the development of modern medical research, fruit extracts of this plant are believed to have numerous beneficial actions on arteriosclerosis, cancer, immunity, and stress (Hu et al. Citation1994; Wang and Xia Citation1996). The therapeutic effect of R. roxburghii fruit extracts could be a result of a combination of various compounds that contain ascorbic acid, polyphenols, and superoxide dismutase (Wu et al. Citation1994; Zhang et al. Citation2001). Besides study on the effective components and action mechanism, a good knowledge of genetics information would contribute to its development and utilization strategy. In this study, we assembled and characterized the complete chloroplast (cp) genome sequence of R. roxburghii based on the Illumina pair-end sequencing data.
Sample of R. roxburghii was collected from Meitan (Guizhou, China; 107°28′15.01″ E, 27°46′51.41″ N), and maintained in Research Center for Medicine & Biology of Zunyi Medical University. Genomic DNA was extracted following the modified CTAB method (Doyle and Doyle Citation1987). The whole-genome sequencing was conducted with 150 bp pair-end reads on the Illumina Hiseq Platform (Illumina, San Diego, CA). About 8 million reads were obtained and kept by mapping to cp genomes of Rosa odorata var. gigantea (KF753637) (Yang et al. Citation2014) using BWA (Li and Durbin Citation2009) and SAMtools (Li et al. Citation2009), then assembled using Velvet (Zerbino and Birney Citation2008). Annotation was performed with Plann (Huang and Cronk Citation2015). The complete cp genome sequence together with gene annotations was submitted to GenBank under the accession numbers of KX768420 for R. roxburghii.
The complete cp genome of R. roxburghii is a double stranded, circular DNA 156,749 bp in length, which contains two inverted repeat (IR) regions of 26,053 bp each separated by a large single copy (LSC) and a small single copy (SSC) region of 85,862 and 18,791 bp, respectively. The genome contained 139 genes, including 88 protein-coding genes (82 PCG species), 39 tRNA genes (32 tRNA species), and eight ribosomal RNA genes (four rRNA species). Most of the gene species occur as a single copy, while 17 gene species occur in double copies, including all rRNA species, seven tRNA species and six PCG species. The overall AT content of R. roxburghii cpDNA is 62.8%, while the corresponding values of the LSC, SSC, and IR regions are 64.8, 68.7, and 57.4%, respectively.
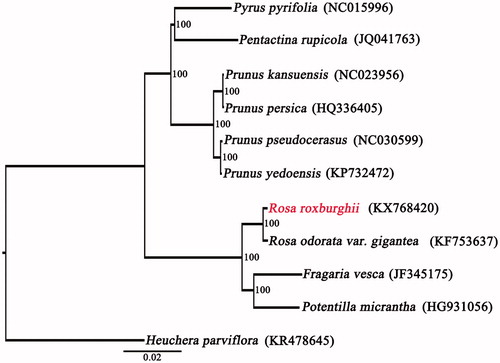
To validate the phylogenetic position of R. roxburghii, a maximum likelihood (ML) tree with 100 bootstrap replicates was inferred using RaxML version 8 (Stamatakis Citation2014) containing plastid genomes of other nine Rosaceae species, Heuchera parviflora was used as the outgroup. The phylogenetic position of R. roxburghii is closely related to R. odorata var. gigantea (). This complete cp genome can be subsequently used for population, phylogenetic and cp genetic engineering studies of R. roxburghii, and such information would be fundamental to formulate potential development and management strategies for this important medicine plant.
Figure 1. Maximum likelihood (ML) tree was constructed with other nine CP genome sequences of Rosaceae, Heuchera parviflora (KR478645) was used as the outgroup. Bootstrap support values (%) are indicated in each node. GenBank accession numbers: Prunus kansuensis (NC023956), P. persica (HQ336405), P. pseudocerasus (NC030599), P. yedoensis (KP732472), Pyrus pyrifolia (NC015996), Pentactina rupicola (JQ041763), Potentilla micrantha (HG931056), Fragaria vesca (JF345175), and Rosa odorata var. gigantea (KF753637).
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hu WY, Bai Y, Han XF, Zheng Q, Zhang HS, He WH. 1994. Anti-atherosclerosis effect of Rosa roxburghii tratt. Chin Pharm J. 25:529–532.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 25:1754–1760.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang W, Xia BN. 1996. Advances in study of Rosa roxburghii tratt. Chin Pharm J. 31:643–646.
- Wu LF, Yang LD, He ZF, Xiong LY, Liang XP, Zhang XB. 1994. Effects of Rosa roxburghii juice on the experimental hyperlipidemia and arteriosclerosis in rabbits. Chin J Vet Sci. 15:386–389.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang CN, Liu XZ, Qiang HJ, LI K, Wang JJ, Chen DN, Zhuang YY. 2001. Inhibitory effects of Rosa roxburghii tratt juice on in vitro oxidative modification of low density lipoprotein and on the macrophage growth and cellular cholesteryl ester accumulation induced by oxidized low density lipoprotein. Clin Chim Acta. 313:37–43.
