Abstract
Genetic analysis is an important tool in understanding population structure, genetic diversity, and phylogenetics of endangered species likely to be affected by microevolution and anthropogenic factors. Western Ghats landscape is one of the identified biodiversity hotspots in India, and micro-evolutionary processes are observed in this landscape due to the presence of the gaps in the mountain ranges. Nilgiri tahr is endemic to and distributed in this landscape while very little is known about genetic characteristics, population structure and impact of these gaps on the species. In the present study, two different populations of Nilgiri tahr from the north (NPG) and south (SPG) of Palghat gap (PG) were studied using the cytochrome b gene (Cyt b; 310 bp) of mtDNA genome in the Western Ghats, India. Two variable sites were observed in the Cyt b fragment while the mean pairwise genetic distance between these two populations was 0.007. All the samples phylogenetically clustered in either north or south of PG. The presence of shallow divergence indicates the presence of suitable habitat in past which may have facilitated movement between NPG and SPG. A subsequent change in Paleo-climatic conditions and gradual formation of PG may have resulted in population diversification during the Pleistocene. Besides, Forensically Informative Nucleotide Sequence (FINS) observed would help in geo-assigning any individual from NPG or SPG to understand the likely influences on population demography due to poaching.
Introduction
Nilgiri tahr is endemic to Western Ghats, India (Schaller Citation1970), and has recently been shifted to the new genus Nilgiritragus hylocrius (Ropiquet and Hassanin Citation2005) from Hemitragus. Based on the molecular phylogeny, Nilgiri tahr is considered the sister group of the Ovis and therefore transferred to a new genus (Ropiquet and Hassanin Citation2005). Most of the Nilgiri tahr populations are fragmented and restricted to the long narrow stretch of 400 km between 11°30′ N to 8°20′ N in the Western Ghats (Schaller Citation1970; Davidar Citation1978). The Nilgiri tahr is a resident of montane grasslands with rocky cliffs at elevations of around 300–2600 m above mean sea level (Abraham et al. Citation2006; Predit et al. Citation2015). The populations of Nilgiri tahr have drastically decreased due to poaching, habitat degradation and other human activity, and therefore assigned as an ‘endangered’ species in the International Union for Conservation of Nature (IUCN) red list as well as in Schedule I under Indian Wildlife (Protection) Act 1972. However, population and conservation status have been studied (Chandran Citation1980; Wilson Citation1980; Rice Citation1984; Abraham et al. Citation2006; Predit et al. Citation2015) and the reasons for local distribution of the Nilgiri tahr are still limited in the areas of Western Ghats due to its preference for a habitat that is predominantly of grasslands. Identifying conservation units (CUs), management units (MUs), and evolutionarily significant units (ESUs) across the range of the species has been a key issue in effective conservation planning of species to retain the genetic diversity. However, no genetic assessment for Nilgiri tahr is available so far except for a study on cross amplification of nuclear markers from the Eravikulam National Park population (Luis et al. Citation2016). Out of the different gaps observed along the Western Ghats, Palghat gap (PG), which is 30–40 km wide, is a major biogeographic barrier for several species (Subramanyam and Nayar Citation1974; Ali and Ripley Citation1987). Recent molecular studies suggest PG is one of the drivers for diversification of various taxa, viz. the Asian elephant, the lion-tailed macaque, bush frogs, and birds e.g. white-bellied shortwing (Vidya et al. Citation2005; Robin et al. Citation2010; Vijayakumar et al. Citation2016) and next to the PG there is another gap present in south of Western Ghats of 7 km wide called Shencottah Gap (Ali and Ripley Citation1987). Therefore, the present study aims to perform a genetic assessment of Nilgiri tahr in the current distribution range of the species in the Western Ghats to identify any divergence between the populations caused by PG. For this, we used cytochrome b (Cyt b) of mtDNA genome to understand phylogeography and genetic diversity of Nilgiri tahr across the north and south of PG. The data on population genetics thus obtained would also help in the field of wildlife forensics for the detection and determination of the poaching hotspots in future as Nilgiri tahr has often been reported to be poached.
Materials and methods
Faecal pellets were collected from North and South of PG from Mukurthi National Park (MNP) and Anamalai Tiger Reserve (ATR) respectively in the Western Ghats (). These pellets were air dried and stored in silica gel covered by clean Kim-wipes (Kimberley-Clark, Irving, TX) to separate the desiccant from the pellets. Genomic DNA from samples was extracted using Qiagen DNeasy Stool DNA extraction kit (Qiagen, Hilden, Germany) as per manufacturer specified protocols with minor modifications where samples were incubated at 60 °C in a dedicated facility for faecal DNA extraction. The quantity and quality of the extracted DNA were estimated on 0.8% agarose gel stained with ethidium bromide. Polymerase chain reaction (PCR) was used to amplify the mitochondrial part of universal Cyt b (381 bp) gene fragment (Helm-Bychowski and Cracraft Citation1993). PCR was performed in a reaction mixture of 25 μl containing, 10 μl 2 × PCR master mix (Thermo Fisher Scientific, Waltham, MA), 4 μl template DNA, 1 μl primers (0.2 μM each), and 10 μl nuclease-free water. The amplification conditions were 94 °C for 5 min followed by 35 cycles at 94 °C for 30 s, 53 °C for 45 s, and 72 °C for 45 s, and a final extension was done at 72 °C for 10 min. The success of PCR amplification was verified using 2% agarose gel electrophoresis. The amplified products were then processed for cycle sequencing PCR using the Big Dye Terminator Cycle Sequencing Kit version 3.1 (Applied Biosystems, Foster City, CA) and were sequenced on an Applied Biosystems ABI 3500 Genetic Analyzer (Foster City, CA).
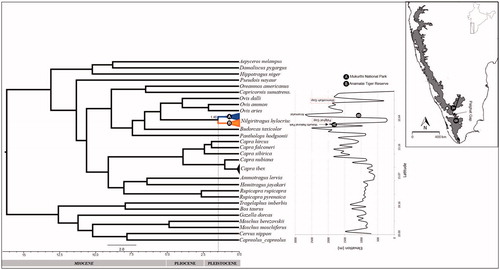
Figure 1. Phylogenetic tree (Bayesian) showing a divergence between two populations (North (A) and South (B) of Palghat Gap) of Nilgiri tahr in the Western Ghats, India (elevation profile adapted from Robin et al. Citation2010).
Data analysis
Each Cyt b sequence was manually scrutinized using SEQUENCHER 4.8 (Gene Codes Corporation, Ann Arbor, MI). Total 310 bp long nucleotides were found suitable for the analysis after validating the sequences. Clean electropherograms and high-quality value (Q > 20) were considered for detecting mutation. Multiple sequence alignments were performed using the CLUSTAL W algorithm implemented in BIOEDIT version 7.0.5.33 (Hall Citation1999). The percent sequence divergence between clusters was calculated using Kimura’s two-parameter (K2P) method in MEGA version 6 (Tamura et al. Citation2013). Phylogenetic trees were reconstructed using different statistical methods, e.g. neighbour-joining (NJ), maximum likelihood (ML), and maximum parsimony (MP) implemented in the MEGA version 6 (Tamura et al. Citation2013). For distance and likelihood-based trees, we used a model of nucleotide substitution estimated in Modeltest 3.6 (Nylander Citation2004), and HKY model was used for the analysis based on the Akaike information criterion (AIC) value. Confidence in the estimated relationships was determined by calculating bootstrap values with 1000 replicates. Bayesian inferences (BIs) were conducted in BEAST v. 2.1.3 (Bouckaert et al. Citation2014). For the molecular clock rate, we used a normal prior and calibrated with the spilt between the Bovidae and Moschidae with mean divergence times of 16 Ma. We placed a standard deviation on the rate equal to 10% of the mean to account for variation and uncertainty in the rate. Dating analyses were performed for 20 million generations while sampling every 1000th tree, and the first 10% of trees sampled were treated as burn-in, and FigTree v.1.3.1 (Rambaut Citation2009) was used to display and summarize annotated phylogenetic trees yielded by BEAST.
Results and discussion
Sequence variability, genetic diversity, and demographic history
A total of 26 samples were sequenced from both the populations and two variable sites were found. Both the point mutations are transition A–G at the position of 14456 and 14472. Within the populations, no variable sites were observed. The mean pairwise genetic distance between the populations was 0.007 which is within the range of intra-species sequence divergence (Tobe et al. Citation2010). The overall nucleotide and haplotype diversity observed were 0.00337 and 0.520 (). Due to lack of within-population variable sites, diversity indices were not obtained within populations. We also compared the haplotype and nucleotide diversity of Nilgiri tahr with other Bovidae species and found a moderate level of nucleotide and haplotype diversity with the different mitochondrial genes (). We compared our data-sets with the different sets of genes in two species showing low genetic diversity comparable to Nilgiri tahr, viz. Gazella arabica acacia (Hd, 0.327; π 0.0051) and Ovis ammon (π 0.0054). The explanation provided for the low genetic diversity in Ovis ammon was recent population divergence (Tserenbataa et al. Citation2004), whereas Gazella arabica acacia has been found to have gone through population bottleneck being restricted to a small area (Hadas et al. Citation2015). In Nilgiri tahr, FST value between the populations (FST=1) suggests there is no sharing of genetic material (). These results have also been supported by the characteristic of two different haplotypes and the fact that no haplotypes have been shared between both the populations of north and south of PG. Both Tajima’s D and Fu’s Fs test were found to be positive () and statistically significant (p<.05) in the combined data. This indicates strong population subdivision, loss of rare alleles or population bottleneck (Hudson et al. Citation1987; Rogers and Harpending Citation1992; Fu Citation1997).
Table 1. Observed diversity and neutrality indices for the Nilgiri tahr population in the Western Ghats, India in comparison with other species of Bovidae.
Phylogenetic, molecular dating, and diversification
In the phylogenetic tree, Nilgiri tahr populations cluster into two reciprocally monophyletic clades, which correspond to north and south of PG (). However, the presence of long branches of Nilgiri tahr divergence from the Ovis probably indicates ancestor of this species was widely distributed across the Western Ghats (). The divergence time estimates using fossils calibration, the point of the split between the Moschidae and Bovidae estimates the divergence between the populations of north and south of PG to be around 1.46 (95% highest posterior density; 0.80–2.34) million year ago (Ma). The presence of two diverse populations in the north and south of PG indicates that this gap may have played some role in population divergence. However, Nilgiri tahr is a less studied animal, and till now no evidence for the population divergence has been reported except the phylogenetic study confirmed Nilgiri tahr as the sister group of Ovis with the divergence time range from Ovis around 2.7–5.2 Ma (Ropiquet and Hassanin Citation2005). However, the origin of PG is debated and dated around 500 Ma ago (Soman et al. Citation1990) and is earlier than the reported divergence time for most of the species in the Western Ghats. Published studies on various taxa reveals that PG has been one of the prominent barriers to gene flow in the Asian elephants (Vidya et al. Citation2005), lion-tailed macaques (Ram et al. Citation2015), birds, e.g. white-bellied shortwing (Brachypteryx major) (Robin et al. Citation2010) and 10 other species (Robin et al. Citation2015), Nyctibatrachidae family of frogs (Van-Bocxlaer et al. Citation2012) and bush frogs (Vijayakumar et al. Citation2016) in the Western Ghats. PG also acts as a barrier for the gene flow in the plant species like Eurya nitida (Bahulikar et al. Citation2004) and Gaultheria fragrantissima (Apte et al. Citation2006). In this context, the divergence time estimated for different species varies from 0.29 Ma in the Asian elephant (Vidya et al. Citation2005), 2.4 Ma for lion-tailed macaques (Ram et al. Citation2015) to 2.5 Ma in the bush frog (Vijayakumar et al. Citation2016) and 40 Ma for the frog species of family Nyctibatrachidae (Van-Bocxlaer et al. Citation2012). Fish species of genus Puntius (red line barb) shows the greater divergence time 56 Ma than these observed for these taxa in north and south of PG (John et al. Citation2013). The divergence time obtained for Nilgiri tahr (1.46 Ma) is older than that observed for the elephant (0.29 Ma) but within the range of lion-tailed macaque and bush frogs (2.4–2.5 Ma).
PG acts as the potential barrier in spite of land connectivity across the 40 km wide gap. However, no extensive survey on palaeobotany is available which may elucidate how this gap acts as the barrier. Moreover, studies on the bush frog (Vijayakumar et al. Citation2016), Caecilians (Gower et al. Citation2007) and the shallow divergence in Nilgiri tahr as discussed in this study do not highlight PG as a significant barrier. In support of this, literature also reveals that PG has gradually formed by the differential uplift of the plateau on both sides and further erosion cycles due to the fluvial actions of westerly flowing streams (Rao and Srinivasan Citation1980; Medlicott and Blanford Citation1983). A detailed intra-specific phylogeographic study of different taxa is needed to identify the cause of the phenomenon (Joshi and Karanth Citation2013). Moreover, high human activity and loss of forest connectivity have also been considered as one of the cause for population differentiation (Vidya et al. Citation2005; Robin et al. Citation2015) though divergence time estimates reported are older than the anthropogenic activities, only supporting the theory of the Pleistocene diversification. Pleistocene climatic oscillations have been suggested to be the cause of the diversification and disjunct distribution of species in different landscapes (Clemens et al. Citation1991; McClymont et al. Citation2013), which is in accordance with the divergence time of the species studied so far across the north and south of PG in the Western Ghats.
Conservation implication
The mitochondrial marker (Cyt b) used in the current study suggests the presence of reciprocally monophyletic clades in Nilgiri populations of north and south of PG in the Western Ghats. The literature reveals that the northern parts have been relatively drier than the southern Western Ghats during the Pleistocene and acted as a refugium for the species (Sukumar et al. Citation1993). Nilgiri and Annamalai regions in the Western Ghats display a phytogeographic barrier to have different floristic compositions (Subramanyam and Nayar Citation1974; Vidya et al. Citation2005). Such spatial differences in plant species and moisture regimes in the habitat are known to result in intra-specific variations in wild herbivores. Kangas et al. (Citation2016) reported the presence of intra-specific geographic variation in the shape of mandible across the range of moose (Alces alces) regarding attachment surfaces of the muscles that control biting and mastication as a response to food plant characteristics. Due to lack of information on such anatomical variations in Nilgiri tahr, we believe that such difference may exist to harvest nutrients differently from plant species in these two populations of dry and moist regions of Western Ghats. Therefore, we suggest both the populations of the Nilgiri tahr should also be managed as a separate MU for in situ conservation plans as well as being considered as separate entities for ex situ conservation strategies. Besides, we also reiterate that thorough studies on anatomical differences, microsatellite and other mitochondrial markers are required to understand the extent of population divergence and gene flow in Nilgiri tahr populations across the Western Ghats.
Acknowledgements
We thank Director, Dean and Research Coordinator, Wildlife Institute of India, India Dehradun for supporting this work. We extend our thanks to Nodal Officer(s), Wildlife Forensic and Conservation Genetic Cell and Wildlife Biology Research Lab for the laboratory support. We also thank Forest Department of Karnataka, Kerala and Tamil Nadu for granting us permission for the samples collection and consistent support in the field collection.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abraham SK, Easa PS, Sivaram M. 2006. Status and distribution of Nilgiri Tahr Hemitragus hylocrius in Kerala part of the Western Ghats. Zoos Print J. 21:2379–2385.
- Ali S, Ripley SD. 1987. Compact handbook of the birds of India and Pakistan: together with those of Bangladesh, Nepal, Bhutan and Sri Lanka. 2nd ed. New Delhi: Oxford University Press.
- Apte GS, Bahulikar RA, Kulkarni RS, Lagu MD, Kulkarni BG, Suresh HS, Rao PSN, Gupta VS. 2006. Genetic diversity analysis in Gaultheria fragrantissima Wall. (Ericaceae) from the two biodiversity hotspots in India using ISSR markers. Curr Sci. 91:1634–1640.
- Bahulikar RA, Lagu MD, Kulkarni BG, Pandit SS, Suresh HS, Rao MKV, Ranjekar PK, Gupta VS. 2004. Genetic diversity among spatially isolated populations of Eurya nitida Korth. (Theaceae) based on inter-simple sequence repeats. Curr Sci. 86:824–831.
- Bouckaert R, Heled J, Khnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10:1–6.
- Chandran PR. 1980. Nilgiri Tahr (Hemitragus hylocrius) in captivity. J Bombay Nat Hist Soc. 77:129–130.
- Clemens S, Prell W, Murray D, Shimmield G, Weedon G. 1991. Forcing mechanisms of the Indian Ocean monsoon. Nature. 354:56–58.
- Davidar ER. 1978. Distribution and status of the Nilgiri tahr (Hemitragus hylocrius)—1975–78. J Bombay Nat Hist Soc. 75:815–844.
- Fu YX. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 147:915–925.
- Gower DJ, Dharne M, Bhatta G, Giri V, Vyas R, Govindappa V, Oommen OV, George J, Shouche Y, Wilkinson M. 2007. Remarkable genetic homogeneity in unstriped, long-tailed Ichthyophis along 1500 km of the Western Ghats, India. J Zool. 272:266–275.
- Hadas L, Hermon D, Boldo A, Arieli G, Gafny R, King R, Bar-Gal GK. 2015. Wild gazelles of the southern levant: genetic profiling defines new conservation priorities. PLoS One. 10:1–18.
- Hall T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Helm-Bychowski K, Cracraft J. 1993. Recovering phylogenetic signal from DNA sequences: relationships within the corvine assemblage (class aves) as inferred from complete sequences of the mitochondrial DNA cytochrome-b gene. Mol Biol Evol. 10:1196–1214.
- Hudson RR, Kreitman M, Aguadé M. 1987. A test of neutral molecular evolution based on nucleotide data. Genetics. 116:153–159.
- John L, Philip S, Dahanukar N, Anvar Ali PH, Tharian J, Raghavan R, Antunes A. 2013. Morphological and genetic evidence for multiple evolutionary distinct lineages in the endangered and commercially exploited red lined torpedo barbs endemic to the Western Ghats of India. PLoS One. 8:e69741.
- Joshi J, Karanth P. 2013. Did southern Western Ghats of peninsular India serve as refugia for its endemic biota during the Cretaceous volcanism? Ecol Evol. 3:3275–3282.
- Kangas V-M, Rytkönen S, Kvist L, Käpylä T, Nygrén T, Aspi J. 2016. Geographic cline in the shape of the moose mandible: indications of an adaptive trend. J Mamm Evol. 24:233–241. http://link.springer.com/10.1007/Q3 s10914-016-9344-yJ
- Liu W, Yao YF, Yu Q, Ni QY, Zhang MW, Yang JD, Mai MM, Xu HL. 2013. Genetic variation and phylogenetic relationship between three serow species of the genus Capricornis based on the complete mitochondrial DNA control region sequences. Mol Biol Rep. 40:6793–6802.
- Luis A, Davidar P, Reddy PA. 2016. Potential of cross-species microsatellite markers to assess population genetics of the endemic, endangered Nilgiri tahr (Nilgiritragus hylocrius). Eur J Wildl Res. 63:16.
- McClymont EL, Sosdian SM, Rosell-Melé A, Rosenthal Y. 2013. Pleistocene sea-surface temperature evolution: early cooling, delayed glacial intensification, and implications for the mid-Pleistocene climate transition. Earth-Sci Rev. 123:173–193.
- Medlicott HB, Blanford WT. 1983. A manual of geology of India. 2nd ed. Calcutta: Wentworth Press.
- Nylander JAA. 2004. Mr Modeltest v2. Program distributed by the author evolutionary biology centre. Uppsala: Uppsala University.
- Predit PP, Prasath V, Raj M, Desai A, Zacharia J, Johnsingh AJ, Ghose D, Ghose P, Sharma RK. 2015. Status and distribution of the Nilgiri tahr in the Western Ghats, India. http://journals.cambridge.org/abstract_S0030605300013442
- Ram MS, Marne M, Gaur A, Kumara HN, Singh M, Kumar A, Umapathy G. 2015. Pre-historic and recent vicariance events shape genetic structure and diversity in endangered lion-tailed macaque in the Western Ghats: implications for conservation. PLoS One. 10:1–16.
- Rambaut A. 2009. FigTree, ver. 1.3.1 available at http://tree.bio.ed.ac.uk/software/figtree/
- Rao JN, Srinivasan R. 1980. Proceedings of the Seminar on Geology and Geomorphology. GS1 Special Publication. p. 41–45.
- Rice CG. 1984. The behaviour and ecology of Nilgiri Tahr (Hemitragus hylocrius Ogilby, 1938) [dissertation]. College Station (TX): A&M Univ. p. 254.
- Robin VV, Sinha A, Ramakrishnan U. 2010. Ancient geographical gaps and paleo-climate shape the phylogeography of an endemic bird in the sky islands of Southern India. PLoS One. 5:e13321.
- Robin VV, Vishnudas CK, Gupta P, Ramakrishnan U. 2015. Deep and wide valleys drive nested phylogeographic patterns across a montane bird community. Proc R Soc Lond B Biol Sci. 282:20150861. http://rspb.royalsocietypublishing.org/content/282/1810/20150861.abstract
- Rogers A, Harpending HM. 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 9:552–569.
- Ropiquet A, Hassanin A. 2005. Molecular evidence for the polyphyly of the genus Hemitragus (Mammalia, Bovidae). Mol Phylogenet Evol. 36:154–168.
- Schaller GB. 1970. Observations on Nilgiri tahr (Hemitragus hylocrius Ogilby, 1838). J Bombay Nat Hist Soc. 67:365–389.
- Soman K, Thara K, Arakelyants M, Golubyev V. 1990. Mineral ages of pegmatites from the Palghat gap region in Kerala and their tectonic significance. J Geol Soc India. 35:82–86.
- Subramanyam K, Nayar M. 1974. Vegetation and phytogeography of the Western Ghats. In: Mani MS, editor. Ecology and biogeography in India. The Hague: Dr W Junk Publishers.
- Sukumar R, Ramesh R, Pant RK, Rajgopalan G, Rajagopalan G. 1993. A d13 C record of late Quaternary climate change from tropical peats in southern India. Nature. 364:703–706.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tan S, Zou D, Tang L, Wang G, Peng Q, Zeng B, Zhang C, Zou F. 2012. Molecular evidence for the subspecific differentiation of blue sheep (Pseudois nayaur) and polyphyletic origin of dwarf blue sheep (Pseudois schaeferi). Genetica. 140:159–167.
- Tanchenge D, Hongjun C, Tong H, Hongpan W, Lei H, Yan G. 2016. Genetic diversity and phylogenetic status of Gazella subgutturosa at the Mountain Kalamaili Ungulate Nature Reserve, Xinjiang. Acta Theriol Sin. 36:77–86.
- Tobe SS, Kitchener AC, Linacre AMT. 2010. Reconstructing mammalian phylogenies: a detailed comparison of the cytochrome b and cytochrome oxidase subunit i mitochondrial genes. PLoS One. 5:e14156.
- Tserenbataa T, Ramey RR, Ryder OA, Quinn TW, Reading RP. 2004. A population genetic comparison of argali sheep (Ovis ammon) in Mongolia using the ND5 gene of mitochondrial DNA; implications for conservation. Mol Ecol. 13:1333–1339.
- Van-Bocxlaer I, Biju SD, Willaert B, Giri VB, Shouche YS, Bossuyt F. 2012. Mountain-associated clade endemism in an ancient frog family (Nyctibatrachidae) on the Indian subcontinent. Mol Phylogenet Evol. 62:839–847.
- Vidya TNC, Fernando P, Melnick DJ, Sukumar R. 2005. Population differentiation within and among Asian elephant (Elephas maximus) populations in southern India. Heredity (Edinb). 94:71–80.
- Vijayakumar SP, Menezes RC, Jayarajan A, Shanker K. 2016. Glaciations, gradients, and geography: multiple drivers of diversification of bush frogs in the Western Ghats Escarpment. Proc R Soc B Biol Sci. 283:20161011.
- Wilson C. 1980. The breeding and management of Nilgiri tahr Hemitragus hylocrius at Memphis Zoo. Int Zoo Yearb. 20:104–106.
- Zeng B, Xu L, Yue B, Li Z, Zou F. 2008. Molecular phylogeography and genetic differentiation of blue sheep Pseudois nayaur szechuanensis and Pseudois schaeferi in China. Mol Phylogenet Evol. 48:387–395.
- Zhang F, Jiang Z, Xu A, Zeng Y, Li C. 2013. Recent geological events and intrinsic behavior influence the population genetic structure of the Chiru and Tibetan Gazelle on the Tibetan Plateau. PLoS One. 8:e60712.
- Zhang F, Jiang Z. 2006. Mitochondrial phylogeography and genetic diversity of Tibetan gazelle (Procapra picticaudata): implications for conservation. Mol Phylogenet Evol. 41:313–321.
