Abstract
This study determines the presence of R. tanezumi from in Nepal using morphological and molecular analyses. Morphologically, it is indistinguishable with R. rattus owing to similar fur colour and morphometric data. However, molecular identification and phylogenetic analysis using sequences of the mitochondrial DNA (mtDNA) Cytochrome B (CytB) gene revealed two different species R. rattus and R. tanezumi from collected specimens. The genetic distance between R. rattus and R. tanezumi was found 0.043. In phylogenetic tree, the clade of R. tanezumi is distinguished into two sub-clades, R. tanezumi found in Nepal, and East Asian countries, China, Laos, Thailand, Viet Nam, and South Korea have genetic distance 0.031, suggesting the different lineages of R. tanezumi. This study confirmed the R. tanezumi present in Nepal. Our findings suggest that morphological analysis and molecular study should be carried out simultaneously for accurate identification of small sized cryptic mammals like R. tanezumi and R. rattus.
Introduction
The Oriental house rat, Rattus tanezumi, is an indigenous species of South East Asia (Niethammer and Martens Citation1975) that has been introduced to East Asia and Africa through transportation by humans (Musser and Carleton Citation2005). Some reviewers have mentioned its presence in Nepal, but they have not provided sufficient evidence for its justification (Pearch Citation2011; Thapa Citation2014). In fact, it is a morphologically indistinguishable species with a sister taxon, R. rattus (Aplin et al. Citation2003a; Musser and Carleton Citation2005). These cryptic species can be differentiated using either cytogenetic or molecular techniques. In karyotype studies, they can be differentiated based on different numbers of chromosomes (Baverstock et al. Citation1983; Chingangbam et al. Citation2014). Meanwhile, in molecular studies, the differentiation can be carried out by analysis of intra-specific genetic divergence using nucleotide sequences including the mitochondrial DNA (mtDNA) Cytochrome B (CytB) gene (Brown and Simpson Citation1981; Aplin et al. Citation2011).
However, previous taxonomic studies on Nepalese Rattus were limited to morphological studies, so there has been continuing confusion regarding morphologically similar and sympatric taxa such as R. tanezumi and R. rattus. In this study, data from morphological and molecular analyses were integrated to distinguish R. tanezumi and R. rattus collected in Nepal.
Materials and methods
Specimen collection was carried out in Lumbini, Pokhara, and Kathmandu, Nepal, from 2014 to 2016 by using Sherman live traps (). Field identification was carried out using external morphology and earlier reports (Ellerman Citation1961; Aplin et al. Citation2003a; Baral and Shah Citation2008). Examination of external morphology included fur colour, footpad, tail, ear morphology, and pairs of mammary glands in females as well as measurement of body weight (BW), head–body length (HBL), tail length (TL), hind foot length (HFL), and ear length (EL). The independent-sample t-test was used to compare the means of morphological characters between the two species R. tanezumi and R. rattus, and one-way analysis of variance (ANOVA) was used to assess the significant differences between the two species, using IBM SPSS 20.0 (IBM Corp. Armonk, NY).
Table 1. Samples used in this study.
The tip of the tail of each individual rat was cut off and kept in a sterile tube for DNA extraction. Total DNA was extracted from the tissue sample using Wizard Genomic DNA Purification Kit (Promega, Wisconsin, MI). MtDNA CytB was amplified using primers L14724 and H15915 designed by Irwin et al. (Citation1991). Polymerase chain reactions (PCRs) were performed according to the procedure of Adhikari et al. (Citation2017). The purified PCR products amplified were directly sequenced with a DNA sequencing ABI 3130XL Genetic Analyzer (Applied Biosystems, Foster, CA). All DNA sequences were subjected to a similarity search using the Basic Local Alignment Search Tool (BLAST) of the National Center for Biotechnology Information (NCBI) database and listed out the most identical putative species.
Multiple sequence alignments were executed using the mtDNA CytB sequences of R. rattus and R. tanezumi determined in this study and reference sequences of Rattus species taken from NCBI database (), which were carried out by using the CLUSTAL W program (Larkin et al. Citation2007). The CytB haplotypes (H) were determined using the DNASP v5 program (Librado and Rozas Citation2009). Genetic distance was calculated between R. rattus and R. tanezumi and intergroup genetic distance was calculated between R. tanezumi recorded in Nepal and East Asian countries China, Laos, Thailand, Viet Nam, and South Korea. Phylogenetic relationships were inferred using maximum likelihood (ML) (Felsenstein Citation1981) and Bayesian inference (BI) (Huelsenbeck and Ronquist Citation2001) based on CytB sequences (). In both methods, the best-fit nucleotide substitution model and parameters were determined using the Akaike information criterion (AIC) (Posada and Buckley Citation2004). In ML analysis, model selection was performed using MEGA 7.0 program (Kumar et al. Citation2016), and the Tamura–Nei model with the gamma distribution (T93 + G) was selected. The ML with bootstrapping (1000 replications) and intergroup genetic distance were also performed using MEGA 7.0 program. In BI, model selection was carried out using MrModeltest 2.3 (Nylander Citation2004), and General Time Reversible model with gamma distribution plus invariant sites (GTR + G + I) was selected. The Bayesian phylogenetic tree was generated using MrBayes 3.2.3 program (Ronquist et al. Citation2012). Four Markov Chain Monte Carlo (MCMC) chains were run for 100,000 generations, sampling every 100 generations, with the first 250 sampled trees discarded as ‘burn-in’ and a 50% majority rule consensus tree was constructed. Reliabilities for inferred nodes were examined with posterior probabilities. Tentative divergence times for all branching points in the ML tree topology were calculated with the RelTime method (Tamura et al. Citation2012) in the MEGA 7.0 program. The minimum and maximum divergence time were used from the fossil-based calibration interval of Rattus and Mus divergence 11–12.3 million years before present, MYBP (Jacobs and Flynn Citation2005).
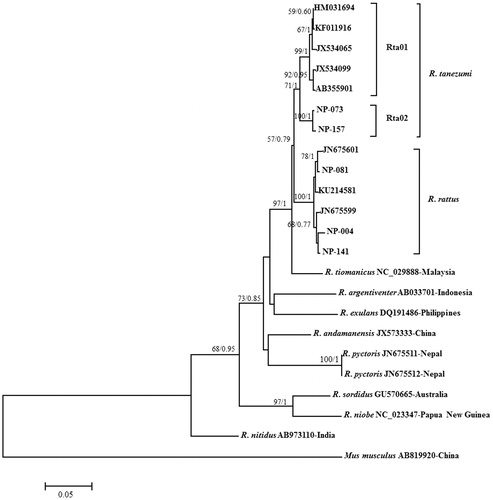
Figure 1. Maximum likelihood and Bayesian tree based on the mtDNA CytB gene sequences for two haplotypes of R. tanezumi and three haplotypes of R. rattus collected from Nepal and reference sequences of various Rattus species taken from NCBI database. Numbers at nodes are support value for the respective clades determined by the methods of ML/BI. CytB sequences of eight Rattus species (R. tiomanicus, R. argentiventer, R. exulans, R. andamanensis, R. pyctoris, R. sordidus, R. niobe and R. nitidus) and Mus musculus were used as outgroups. Detail information of haplotypes corresponding to those in figure is explained in .
Results and discussion
Altogether, 23 individuals of Rattus species (R. tanezumi, n = 8; R. rattus, n = 15) were collected and identified from five study sites in Pokhara and Lumbini of Nepal (). Usually, murid rodents have different colours on their back and belly, but in R. tanezumi and R. Rattus, we could not detect any noticeable colour difference within and between the species, similar to the report by Mostert (Citation2009). Both species have spiky, brownish, greyish to reddish fur on the dorsal surface and uniform greyish to whitish fur on the ventral surface. The colour variation within and between R. tanezumi and R. rattus could be due to variation in collection season, habitat and location. Morphometric measurement and comparison are the key criteria for distinguishing many species of Rattus (Aplin et al. Citation2003a). In adult individuals, average TL was longer in R. tanezumi (194.00 ± 14.50 mm) than in R. rattus (186.35 ± 16.47 mm), but the average values of HBL and BW were lower in R. tanezumi (HBL = 155.00 ± 10.00 mm, BW= 89.50 ± 18.77 g) than in R. rattus (165.75 ± 16.48 mm, BW = 99.50 ± 27.31 g). However, statistically, there was no significant difference between their morphological characters (ANOVA, df = 18, p > .05). Because their morphological characters are indistinguishable, both species have been regarded as the part of R. rattus complex (Aplin et al. Citation2003a; Musser and Carleton Citation2005; Robins et al. Citation2007).
All the CytB gene sequences of collected specimens were subjected to similarity search, which revealed 15 sequences were 99% identical with R. rattus and eight sequences were above 97% identical with R. tanezumi. Altogether, five haplotypes of R. tanezumi and R. rattus were found in the 23 CytB sequences obtained in this study (). Two distinct haplotypes (NP-073 and NP-157) were found in R. tanezumi, collected from Lumbini and three distinct haplotypes (NP-004, NP-081 and NP-141) were found in R. rattus collected from Lumbini and Pokhara. The ML and Bayesian trees were determined using those haplotypes, which produced a robust and identical phylogenetic tree clustered distinctly into two different clades (). Two haplotypes of R. tanezumi were clustered together in the clade of R. tanezumi reported from China (HM031694), Laos (JX534065), Thailand (JX534099), Viet Nam (JQ823462, AB355901), and South Korea (KF011916). However, three haplotypes of R. rattus were clustered in the clade of R. rattus reported from Nepal (KU214581 and JN675599) and Pakistan (JN675601). The genetic distance between R. rattus and R. tanezumi was found 0.043, which was lower than the estimation of Tollenaere et al. (Citation2010). Furthermore, the R. tanezumi clade is divided into two sub-clades, with separate groups of Nepalese specimens (Rt01) and those from Central and East Asian countries China, Laos, South Korea, Thailand, and Viet Nam (Rt02). The inter-group genetic distance between two groups Rt01 and Rt02 was calculated 0.031, which was higher than 0.020 suggested the different lineages (Hubert and Hanner Citation2015). Aplin et al. (Citation2003b) reported that R. tanezumi can be divided into two taxa: one taxon endemic to South East Asia that is abundant in Viet Nam, Laos, and Cambodia and a South Asian taxon abundant in Bangladesh, Northern Viet Nam, and Hong Kong. This study shows that the specimens found in Nepal could be South Asian taxon possibly distributed in Indian continent. It was collected only in low altitude of Nepal, therefore, further study required for generating detailed information regarding the distribution of R. tanezumi in Nepal and surrounding countries. The tentative divergence time between these two groups of R. tanezumi was estimated 0.39 MYBP, indicating that these two groups have recent divergence.
In Nepal, there have not been authentic reports of the presence of R. tanezumi before this study. Musser and Carleton (Citation2005) have mentioned synonyms of R. tanezumi based on the findings of Hodgson (Citation1845) (Mus brunneus and Mus brunneusculus), but a report by Hinton and Fry (Citation1923) argued that those are the subspecies of R. rattus rather than of R. tanezumi. Our morphological and molecular data resolved the taxonomic controversy of R. tanezumi and confirmed its presence in Nepal. It has sympatric association with R. rattus and mostly inhabits human settlements and agricultural land. This study will be significant for the government and mammalogists of Nepal to understand the taxonomy and ecology of R. tanezumi and R. rattus. Our study suggested that an approach integrating morphological and molecular analyses could be appropriate for the accurate and effective identification of cryptic species like R. tanezumi and R. rattus.
Acknowledgements
Authors are grateful to the Department of Forestry, Government of Nepal, for providing the research permission.
Disclosure statement
The authors declare that they have no conflict of interest.
References
- Adhikari P, Han SH, Kim YK, Kim TW, Thapa TB, Subedi N, Adhikari P, Oh HS. 2017. First molecular evidence of Mus musculus bactrianus in Nepal inferred from the mitochondrial DNA Cytochrome B gene sequences. Mitochondr DNA Part A. DOI:10.1080/24701394.2017.1320994
- Aplin KP, Brown PR, Jacob J, Krebs CJ, Singleton GR. 2003a. Field methods for rodent studies in Asia and the Indo-Pacific. Melbourne: BPA Print Group.
- Aplin KP, Chesser T, Have JT. 2003b. Evolutionary biology of the genus Rattus: profile of an archetypal rodent pest. In: Singleton GR, Hinds LA, Krebs J, Spratt DM, editors. Rats, mice and people: rodent biology and management. ACIAR Monograph 96. Canberra: Clarus Design; p. 487–498.
- Aplin KP, Suzuki H, Chinen AA, Chesser RT, Have JT, Donnellan SC, Austin J, Frost A, Gonzalez JP, Herbreteau V, et al. 2011. Multiple geographic origins of commensalism and complex dispersal history of black rats. PLoS One. 6:e26357.
- Baral HS, Shah KB. 2008. Wild mammals of Nepal: rats and mice. Kathmandu: Himalayan Nature. p. 78–90.
- Baverstock PR, Adams M, Maxson LR, Yosida TH. 1983. Genetic differentiation among karyotypic forms of the black rat, Rattus rattus. Genetics. 105:969–983.
- Brown GG, Simpson MV. 1981. Intra- and interspecific variation of the mitochondrial genome in Rattus norvegicus and Rattus rattus: restriction enzyme analysis of variant mitochondrial DNA molecules and their evolutionary relationships. Genetics. 97:125–143.
- Chingangbam DS, Laishram JM, Singh NB, Taibangjam L, Brajakishore C. 2014. Karyotype evolution and species differentiation in the genus Rattus of Manipur, India. Afr J Biotechnol. 13:4733–4744.
- Ellerman JR. 1961. Rodentia. In: Roonwal ML, editor. The fauna of India including Pakistan, Burma and Ceylon. 2nd ed. Calcutta: Baptist Mission Press; p. 485–884.
- Felsenstein J. 1981. Evolutionary trees from DNA sequences- a maximum-likelihood approach. Confidence limits on phylogenies: an approach using the bootstrap. J Mol Evol. 17:368–376.
- Hinton MAC, Fry TB. 1923. Report No. 37: Nepal. Bombay Natural History Society’s mammal survey of India, Burma and Ceylon. J Bomb Nat Hist Soc. 29:399–428.
- Hodgson BH. 1845. On the rats, mice, and shrews of the central region of Nepal. Ann Mag Nat Hist. 15:266–270.
- Hubert N, Hanner R. 2015. DNA barcoding, species delineation and taxonomy: a historical perspective. DNA Barcodes. 3:44–58.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Irwin DM, Kocher TD, Wilson AC. 1991. Evolution of the cytochrome b gene of mammals. J Mol Evol. 32:128–144.
- Jacobs LL, Flynn LJ. 2005. Of mice… again: the Siwalik rodent record, murine distribution, and molecular clocks. In: Lieberman D, Smith R, Kelley J, editors. Interpreting the past: essays on human, primate and mammal evolution. Leiden: Brill Academic Publisher; p. 63–80.
- Jansa SA, Barker FK, Heaney LR. 2006. The pattern and timing of diversification of Philippine endemic rodents: evidence from mitochondrial and nuclear gene sequences. Syst Biol. 55:73–88.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948.
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452.
- Lu L, Chesters D, Zhang W, Li G, Ma Y, Ma H, Song X, Wu H, Meng F, Zhu C, et al. 2012. Small mammal investigation in spotted fever focus with DNA-barcoding and taxonomic implications on rodents species from Hainan of China. PLoS One. 7:e43479.
- Mostert ME. 2009. Molecular and morphological assessment of invasive, inland Rattus (Rodentia: Muridae) congenerics in South Africa and their reservoir host potential with respect to Helicobacter and Bartonella [M.Sc. Thesis]. Pretoria, South Africa: University of Pretoria.
- Musser GG, Carleton MD. 2005. Super family Muridae. In: Wilson DE, Reeder DM, editors. Mammal species of the world. 3rd ed. Baltimore (MA): Johns Hopkins University Press; p. 894–1531.
- Niethammer J, Martens J. 1975. Die Gattungen Rattus and Maxomys in Afghanistan and Nepal. Zeitsch Fur Saugetier. 40:325–355.
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- Pages M, Bazin E, Galan M, Chaval Y, Claude J, Herbreteau V, Michaux J, Piry S, Morand S, Cosson JF. 2013. Cytonuclear discordance among Southeast Asian black rats (Rattus rattus complex). Mol Ecol. 22:1019–1034.
- Pearch MJ. 2011. A review of the biological diversity and distribution of small mammal taxa in the terrestrial ecoregions and protected areas of Nepal. Zootaxa. 3072:1–286.
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 53:793–808.
- Robins JH, Hingston M, Matisoo-Smith E, Ross HA. 2007. Identifying Rattus species using mitochondrial DNA. Mol Ecol Notes. 7:717–729.
- Robins JH, McLenachan PA, Phillips MJ, McComish BJ, Matisoo-Smith E, Ross HA. 2010. Evolutionary relationships and divergence times among the native rats of Australia. BMC Evol Biol. 10:375.
- Ronquist F, Teslenko M, Mark PV, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Suzuki H, Tsuchiya K, Takezaki N. 2000. A molecular phylogenetic framework for the Ryukyu endemic rodents Tokudaia osimensis and Diplothrix legata. Mol Phylogenet Evol. 15:15–24.
- Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S. 2012. Estimating divergence times in large molecular phylogenies. Proc Natl Acad Sci U S A. 109:19333–19338.
- Thapa S. 2014. A checklist of mammals of Nepal. J Threat Taxa. 6:6061–6072.
- Tollenaere C, Brouat C, Duplantier JM, Rahalison L, Rahelinirina S, Pascal M, Mone H, Mouahid G, Leirs H, Cosson JF. 2010. Phylogeography of the introduced species Rattus rattus in the western Indian Ocean, with special emphasis on the colonization history of Madagascar. J Biogeogr. 37:398–410.
- Truong TT, Yoshimatsu K, Araki K, Lee BH, Nakamura I, Endo R, Shimizu K, Yasuda SP, Koma T, Taruishi M, et al. 2009. Molecular epidemiological and serological studies of hantavirus infection in northern Vietnam. J Vet Med Sci. 71:1357–1363.
