Abstract
The complete mitochondrial genome of Epinephelus chlorostigma has been determined. It contains a typically conserved structure including 13 protein-coding genes, 22 tRNA genes, two rRNA genes and one control region, and the whole sequence was 16,894 bp in length. The overall base composition is A 28.70%, C 27.99%, G 16.28%, T 27.03%. Except ND6 and eight tRNA genes, all other mitochondrial genes are encoded on the heavy strand. Phylogenetic tree was constructed based on 12 protein-coding genes sequences of 18 Epinephelus species, four species belong to Cephalopholis as outgroup, the result showed that E. chlorostigma is most closely related to Epinephelus areolatus. We sequenced the complete mitochondrial genome of E. chlorostigma to enrich the resource of molecular markers for examination of phylogenetic relationships in Epinephelus.
Groupers live in reefs or gravel in the temperate and tropical seas. There are over 100 species of grouper in the world, 90% of which are in Asian sea areas. The Brown-spotted grouper, Epinephelus chlorostigma, is one of these valuable varieties which distributed in the Indian Pacific Ocean, it belongs to family Serranidae, genus Epinephelus. In order to find new DNA markers for the future research of population genetics and phylogenetics and taxology, we determined the complete mitogenome of E. chlorostigma (GenBank accession number no. MG739436) by PCR amplification and primer walking sequence method, it would be useful for further understanding the evolution of ratite and conservation genetics of Epinephelus.
In this study, we reported the complete mitogenome of E. chlorostigma from the South China Sea (18°3′35″N 109°24′19″E). Samples stored in a refrigerator of −80 °C with accession number 20171017EC01. The complete mitochondrial genome of E. chlorostigma, with 16,894 bp in length, includes 13 protein-coding genes, 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes and 1 control region (D-Loop). Genes encoding on the genome are similar among all Perciformes (Liu et al. Citation2016; Ayala et al. Citation2017; Kim et al. Citation2017), with the exception of ND6 and eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, Pro), all other mitochondrial genes are encoded on the H-strand (Liu et al. Citation2016). The total length of the 13 protein-coding genes was 11,427 bp, which corresponded to 67.64% of the whole mitochondrial genome. The overall base composition is A 28.70%, C 27.99%, G 16.28%, T 27.03%. The A + T content (56.69%) is higher than G + C content (43.31%), in common with other Perciformes mitogenomes (Bian et al. Citation2016; Chen et al. Citation2016; Ayala et al. Citation2017; Shi et al. Citation2018). Eleven protein-coding genes start with ATG except COX1 with GTG and ATP6 with ATA. For the stop codon, eight protein-coding genes stop with TAA except ND5 with TAG, COX2, ND3, ND4 and CYTB with an incomplete T. The 22 tRNA genes vary from 69 to 76 bp in length. The 12S rRNA is located between tRNA-Phe and tRNA-Val genes is 953 bp in length (Kim et al. Citation2017). The 16S rRNA is located between tRNA-Val and tRNA-Leu genes (Song et al. Citation2016) and is 986 bp in length. The OL (origin of L-strand replication) is located between tRNA-Asn and tRNA-Cys, the same with other Perciformes (Guo et al. Citation2016), and is 43 bp in length. The control region was 1090 bp in length, localized between tRNA-Pro (TGG) and tRNA-Phe (GAA) genes.
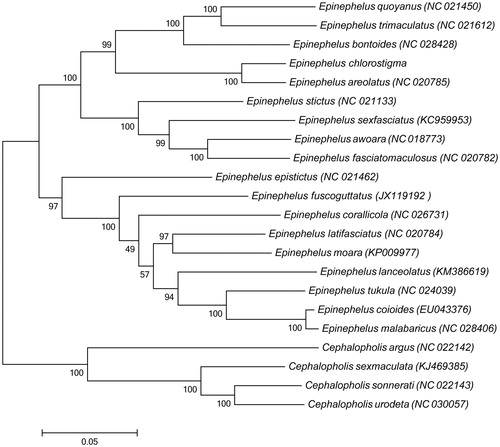
Sequence alignment was conducted by BioEdit (Hall Citation1999). And phylogenetic tree was constructed using maximum-likelihood (ML) method based on the 12 protein-coding genes (except ND6 gene) of 18 available representative species in Epinephelus and four species belong to Cephalopholis as an outgroup (). The result shows that E. chlorostigma is most closely related to Epinephelus areolatus. We expect the present results will provide an important data set for phylogenetic and taxonomic analyses of genus Epinephelus species.
Figure 1. Phylogenetic tree was constructed based on 12 protein-coding genes (except ND6 gene) of 18 Epinephelus complete mitogenome. The black dot indicated the species in this study. The number at each node is the bootstrap probability. The number before the species name is the GenBank accession number.
Disclosure statement
The authors confirm this article content has no conflict of interest, and all the authors are responsible for the content of this article.
Additional information
Funding
References
- Ayala L, Becerra J, Boo GH, Calderon D, DiMarco A, Garcia A, Gonzales R, Hughey JR, Jimenez GA, Jimenez TA, et al. 2017. The complete mitochondrial genome of Xiphister atropurpureus (Perciformes: Stichaeidae). Mitochondrial DNA Part B. 2:161–162.
- Bian L, Su Y, Gaffney PM. 2016. The complete mitochondrial genome sequence of white perch Morone americana (Perciformes, Moronidae). Mitochondrial DNA Part B. 1:380–382.
- Chen H, Deng S, Yang H, Ma X, Zhu C, Huang H, Li G. 2016. Characterization of the complete mitochondrial genome of Priacanthus tayenus (Perciformes: Priacanthidae) with phylogenetic consideration. Mitochondrial DNA Part B. 1:243–244.
- Guo M, Huang H, Gao Y. 2016. Complete mitochondrial genome of darkfin hind Cephalopholis urodeta (Perciformes, Epinephelidae). Mitochondrial DNA Part B. 1:913–916.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Kim YK, Song YS, Kim I-H, Kang C-B, Kim WB, Kim S-Y. 2017. Complete mitochondrial genome of Lycodes tanakae (Perciformes: Zoarcidae). Mitochondrial DNA Part B. 1:947–948.
- Liu Y, Yang M, Zhou T, Xing H, Chen L, Zhang D. 2016. Complete mitochondrial genome of the Antarctic cod icefish, Pagothenia borchgrevinki (Perciformes: Nototheniidae). Mitochondrial DNA Part B. 1:432–433.
- Shi W, Chen S, Yu H. 2018. The complete mitochondrial genome sequence of Siganus sutor (Perciformes: Siganidae). Mitochondrial DNA Part B. 3:62–63.
- Song HY, Hyun YS, Yoon M, Woo J, Lim BJ, Kim HJ, An HS. 2016. The complete mitochondrial genome of Gymnogobius heptacanthus (Perciformes, gobiidae). Mitochondrial DNA Part B. 1:833–834.
