Abstract
Fallopia multiflora is an important Oriental herb belonging to the family Polygonaceae. The F. multiflora mitochondrial genome consists of two circular chromosomes that were 200,352- and 112,098-nucleotides long. The mitochondrial genome encodes 57 genes, including 34 protein-coding genes, 20 transfer RNA (tRNA) genes, and three ribosomal RNA (rRNA) genes. When accounting for 3 overlapping genes, 38 genes were found in chromosome I, and 22 in chromosome II. The phylogenetic analysis suggests that F. multiflora is closely related to Beta macrocarpa and Silene latifolia.
Fallopia multiflora (Hashuoh in Korean) is an important herb in Korea, Japan, and China (Han et al. Citation2016). We extracted genomic DNA from fresh leaves collected from the Oriental Medicine Resource Center of Wonkwang University (geographic coordinate: N 35°56′38″, E 126°57′16″). The whole body specimen was registered to the National Agrobiodiversity Center (http://genebank.rda.go.kr/) under the voucher number IT 272664. A total of 4.76 Gbp of data was obtained using the Illumina MiSeq platform (San Diego, CA). DNA libraries were constructed from the Illumina paired-end (PE) library using the Illumina platform.
The mitochondrial genome of F. multiflora consists of two circular chromosomes, I (GenBank accession number: MF611850), and II (MF611851). The chromosomes were numbered based on the genome size, and the lengths of the chromosomes were 200,352 and 112,098 nucleotides. Fallopia multiflora had two mitochondrial genomes. Multipartite mitochondrial genomes have been reported in some plants, animals, and fungi (Wei et al. Citation2012).
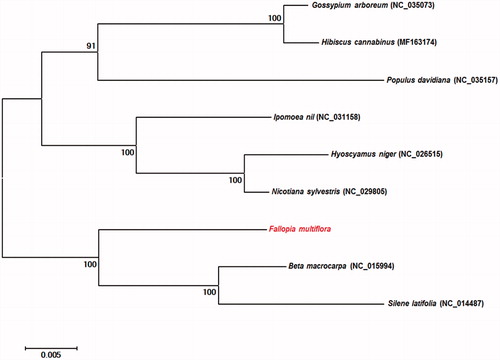
The mitochondrial genome of F. multiflora encoded 57 genes, including 34 protein-coding genes, 20 transfer RNA (tRNA) genes, and three ribosomal RNA (rRNA) genes. Of these 57 genes, 38 were found in chromosome I, and 22 in chromosome II. Three genes overlapped. In addition, we identified 15 open reading frames (ORFs) and splicing variants of the nad1, nad2, and nad5 genes. Interestingly, the nad1 and nad5 splicing genes were located at exons 1, 2, 3 in chromosome II and exons 4, 5 in chromosome I. Phylogenetic relationships of 15 common F. multiflora protein-coding sequences were revealed by comparing data with the genomes of eight other reported species in the Pentapetalae subdivision. The phylogenetic analysis suggested that F. multiflora is closely related to the Beta macrocarpa and Silene latifolia groups ().
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Han EH, Cho K, Goo Y, Kim M, Shin YW, Kim YH, Lee SW. 2016. Development of molecular markers, based on chloroplast and ribosomal DNA regions, to discriminate three popular medicinal plant species, Cynanchum wilfordii, Cynanchum auriculatum, and Polygonum multiflorum. Mol Biol Rep. 43:323–332.
- Wei DD, Shao R, Yuan ML, Dou W, Barker SC, Wang JJ. 2012. The multipartite mitochondrial genome of Liposcelis bostrychophila: insights into the evolution of mitochondrial genomes in bilateral animals. PLoS One. 7:e33973.